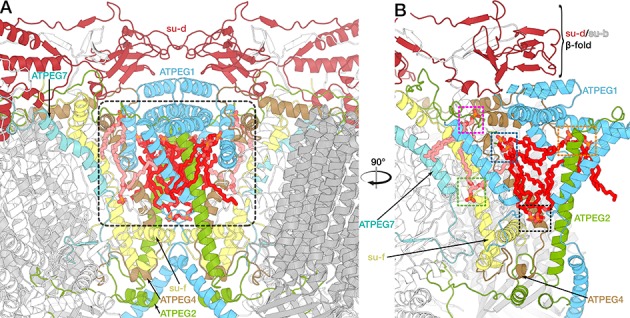
Figure 4. The dimer interface and associated lipids.
(A and B) Views of the dimer interface along (A) and perpendicular (B) to the membrane plane. The dimerisation motifs (interacting subunits coloured) are stacked along the C2-symmetry axis and formed by two copies of subunit d (red) and ATPEG1 (blue), which interacts with its symmetry-related copy, as well as ATPEG2 (green) and subunit f (yellow). Asterisks in (B) indicate positions of lipid-binding sites. (C to G) Close-ups of the lipid-binding sites indicated in (B). Bound lipids at the dimer interface identified as cardiolipin (CDL; C to E) or phospholipids modelled as phosphatidic acid (PL; F and G). Interacting residues (subunits coloured) include at least one arginine residue. Density shown as grey mesh.
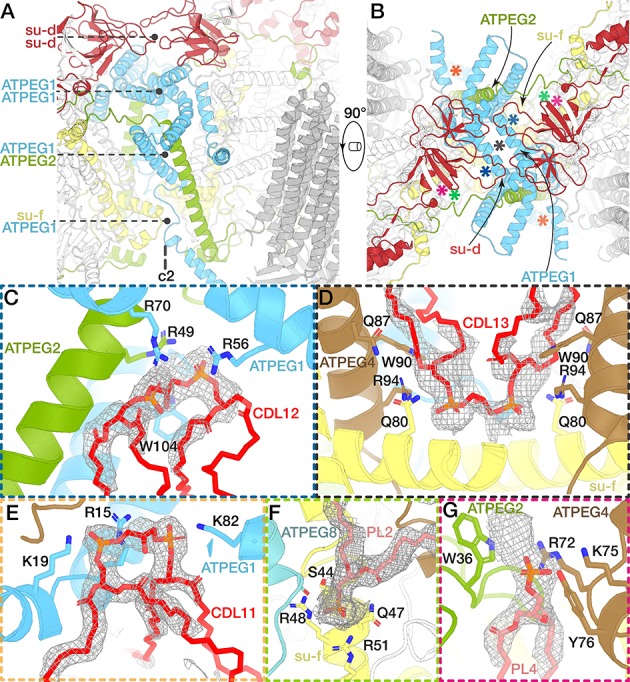
Figure 4—figure supplement 1. Architecture and dimer interface comparison between the yeast and E. gracilis ATP synthase.
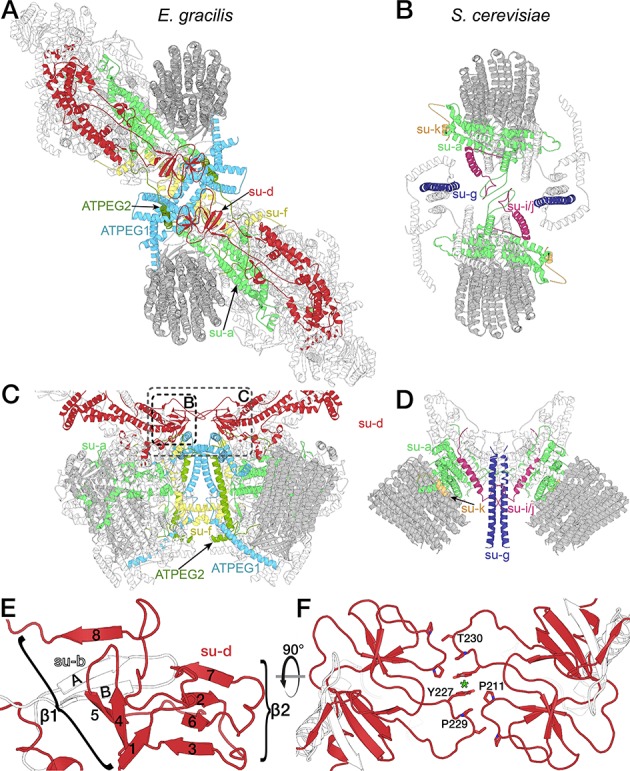
Figure 4—figure supplement 2. Species-specific subunits and extensions form the dimer interface.
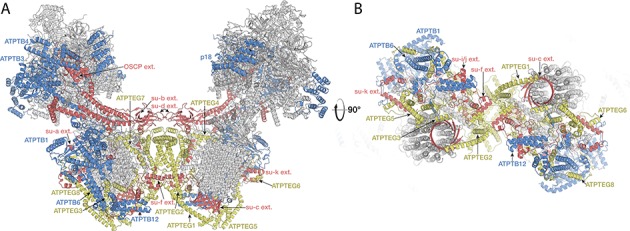
Figure 4—figure supplement 3. Bound lipids of the dimer interface.