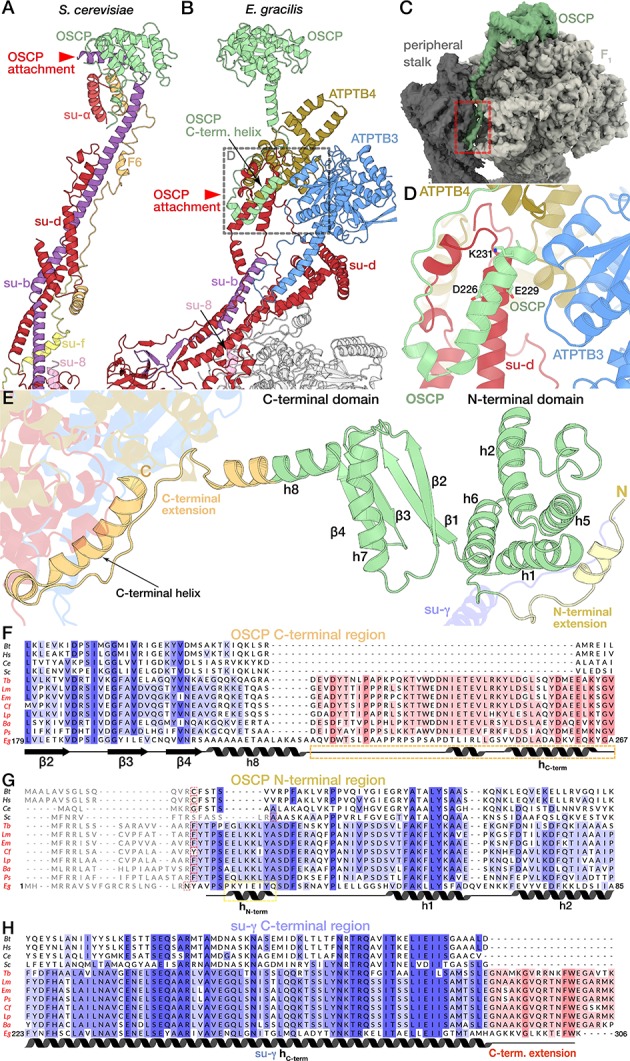
Figure 6. Peripheral Fo subcomplex and the peripheral stalk.
(A) Euglenozoa-specific subunits form a peripheral Fo subcomplex. Density of the Fo with proteins of the peripheral region coloured, c-ring model shown in grey, outline of the detergent belt (yellow dashed lines) with 2 nm offset towards the lumen indicated as determined by the density (transparent gold) (B) Atomic model of the Fo periphery, cavity lipids are shown in magenta. (C to E) Attachment of the peripheral stalk to F1. (C) E. gracilis ATP synthase with proteins constituting the peripheral stalk coloured. (D) Side view of the peripheral stalk tip and F1 (white, crown domain light grey). C-terminal helix of OSCP (light green) extending towards the membrane attaches OSCP to the rest of the peripheral stalk via subunit d (red). (E) The N-terminal extension of OSCP (yellow) interacts with the C-terminal extension of the rotor subunit γ (conserved region purple, extension dark blue).
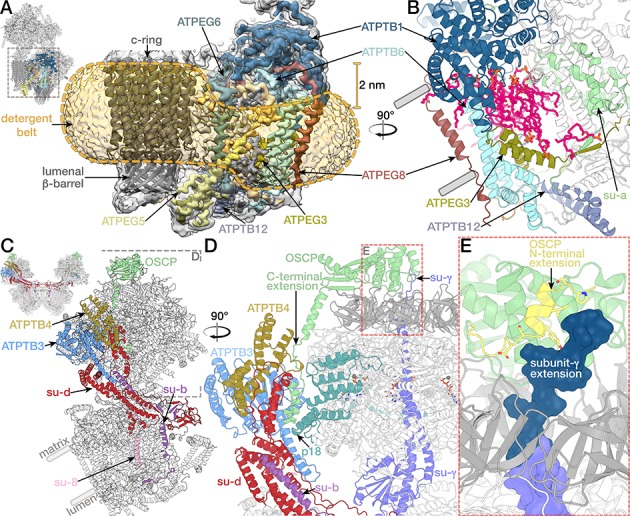
Figure 6—figure supplement 1. The peripheral Fo subcomplex and inverted topology structural motif.
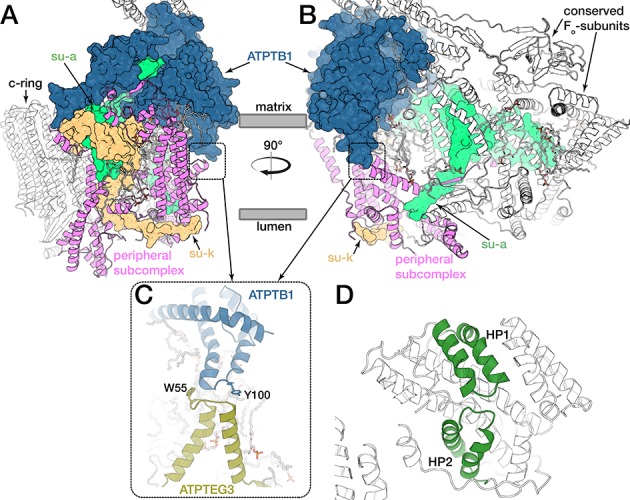
Figure 6—figure supplement 2. Coarse-grained molecular dynamics simulations of the E. gracilis ATP synthase dimer.
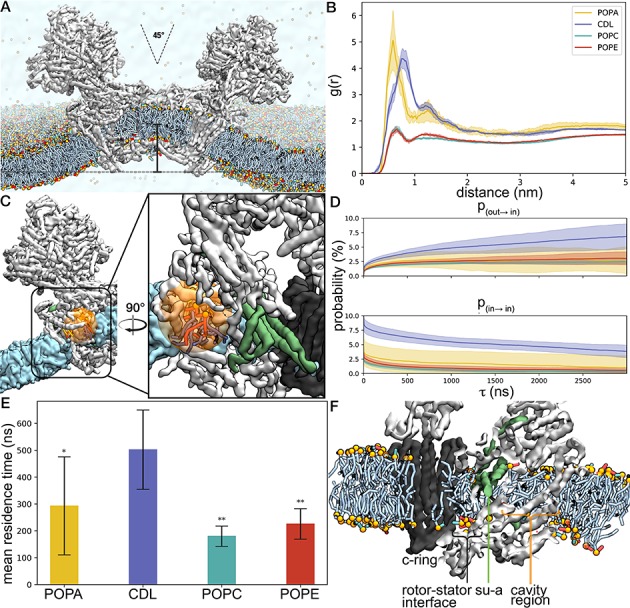
Figure 6—figure supplement 3. Interactions of OSCP extension with F1 and the peripheral stalk.