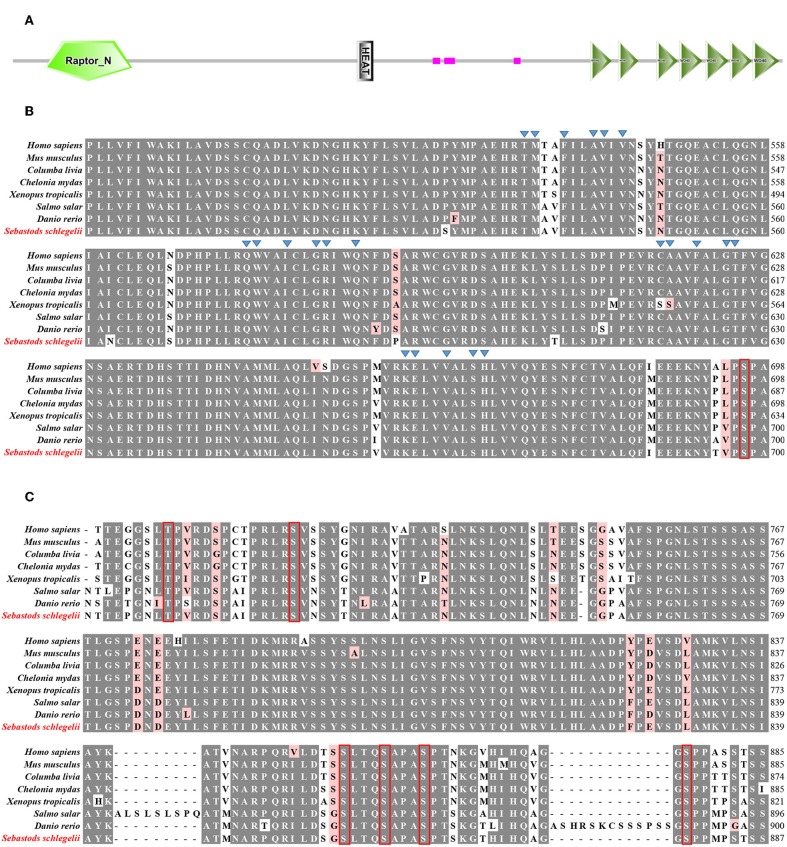
Figure 1.
Sequence alignment of Ss-Raptor with its homologs from other species. (A) The structure of Raptor protein in rockfish by SMART prediction. (B) Multiple sequence alignment analyses of Heat repeat domain in Raptor. Peptide binding sites are indicated with triangle, and the conservative phosphorylation point is shown in red box. (C) Multiple sequence alignment analyses of phosphorylation sites between Heat repeat domain and WD40 domain of Raptor. The conserved phosphorylation point is shown in red box. The background of the amino acid residues conserved in 80% are shown as dark gray and the similar residues is light red. The selected protein sequences are Homo sapiens (NP_065812.1), Mus musculus (NP_065812.1), Columba livia (PKK18984.1), Chelonia mydas (XP_007056109.1), Xenopus tropicalis (XP_012827010.1), Salmo salar (XP_014032262.1), Danio rerio (XP_021327417.1).