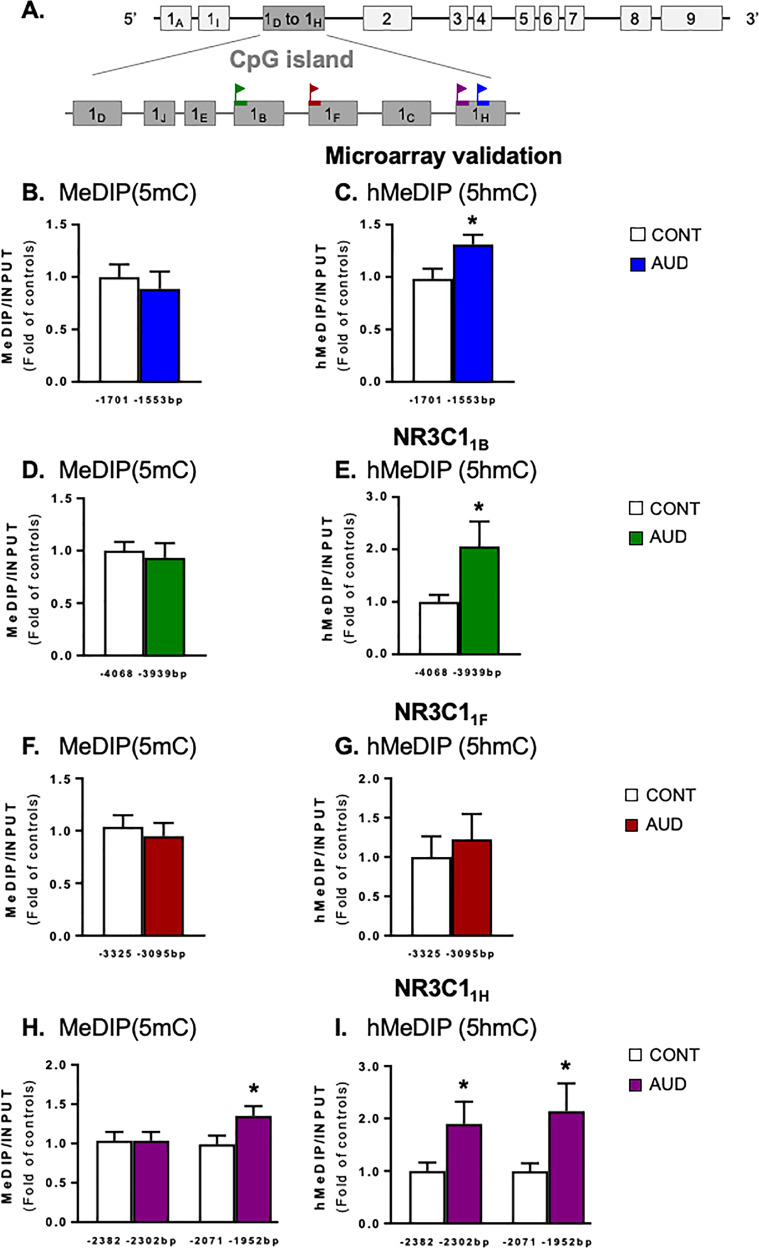
Fig. 2.
Methylation status of NR3C1 exon 1 in the prefrontal cortex (BA10) of alcohol use disorder (AUD) subjects. a Human NR3C1 gene structure. Each rectangle is an exon; the CpG island is illustrated as dark gray rectangles. Flag signs indicate the regions of the gene amplified after (hydroxy)methylated DNA immunoprecipitation [(h)MeDIP] assay or chromatin immunoprecipitation (ChIP) in the untranslated alternative first exons. NR3C1 microarray locus was validated by measuring b methylation and c hydroxymethylation (t1,44 = 2.46, p = 0.018) levels of a region including the CpG detected in the microarray. NR3C1B d methylation and e hydroxymethylation (t1,44 = 2.16, p = 0.036) levels. NR3C1F f methylation and g hydroxymethylation levels. NR3C1H h methylation (from –2071 to –1952 bp, t1,44 = 2.16, p = 0.036) and i hydroxymethylation levels (from –2382 to –2302 bp: t1,44 = 2.01, p = 0.050; from –2071 to –1952 bp: t1,44 = 2.02, p = 0.048). Values are mean±SEM of 23 samples per group for (h)MeDIP assay, 25 controls, and 24 AUD subjects for ChIP. *p < 0.05 Student’s t-test vs. controls