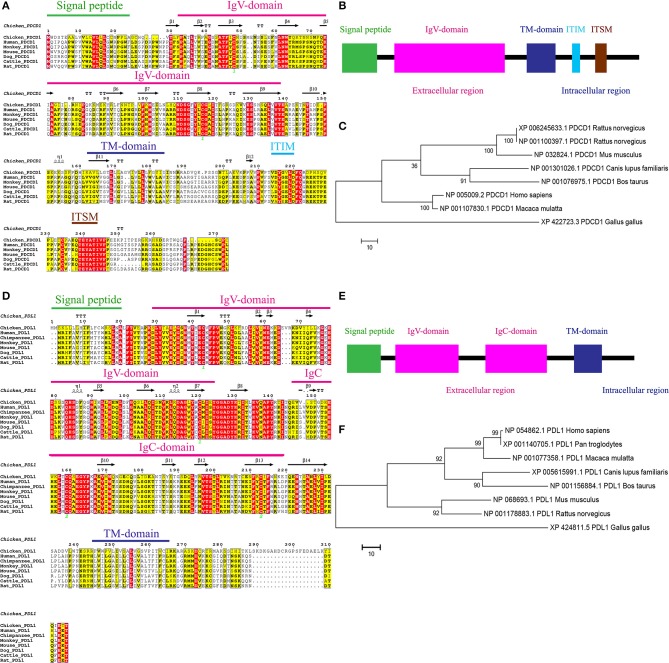
Figure 1.
Alignment of the amino acid sequences of chPD-1 and chPD-L1 with their mammalian orthologs. (A,D) Qualitative analysis of sequence identity and similarity using the ESPript 3.0 online tool. The predicted secondary structures are marked above the alignment (by helices with squiggles, β strands with arrows, and turns with TT letters) and are based on the PD-1 and PD-L1 structural models. Strictly conserved residues are boxed in white on a red background, more conserved residues are boxed in black on a yellow background, and less conserved residues are boxed in black on a white background. Shaded areas represent conservation of amino acid, the darker the shading, the more conserved the residue across species. The green number in Ig V domain of chPD-1 and Ig V and Ig C domains of chPD-L1 indicate that the two cysteine residues that form an intrachain disulfide bridge, respectively. (B,E) Predicted functional motifs in chPD-1 contains extracellular, transmembrane and intracellular domains and chPD-L1 contains extracellular, transmembrane and intracellular domains. (C,F) Maximum likelihood phylogenetic trees based on amino acid sequences of chPD-1 and chPD-L1 in relation to other animal species. Bootstrap values of 1,000 replicates was assigned for the analysis.