Abstract
CRISPR–Cas systems protect prokaryotic cells from invading phages and plasmids by recognizing and cleaving foreign nucleic acid sequences specified by CRISPR RNA spacer sequences. Several CRISPR–Cas systems have been widely used as tool for genetic engineering. In DNA-targeting CRISPR–Cas nucleoprotein effector complexes, the CRISPR RNA forms a hybrid with the complementary strand of foreign DNA, displacing the noncomplementary strand to form an R-loop. The DNA interrogation and R-loop formation involve several distinct steps the molecular details of which are not fully understood. This chapter describes a recently developed fluorometric Cas beacon assay that may be used for measuring of specific affinity of various CRISPR–Cas complexes for unlabeled target DNA and model DNA probes. The Cas beacon approach also can provide a sensitive method for monitoring the kinetics of assembly of CRISPR–Cas complexes.
1. Introduction
CRISPR (clustered regularly interspaced short palindromic repeats)–Cas (CRISPR-associated genes) systems provide prokaryotes with adaptive hereditable immunity by acquiring fragments of DNA (spacers) matching foreign nucleic acid and upon afterward destroying DNA or RNA molecules harboring sequences complementary to acquired spacers (Barrangou et al., 2007; Brouns et al., 2008; Garneau et al., 2010). Based on structural and functional features, the CRISPR–Cas systems have been classified into two classes (1 and 2), six types (I–VI) and many subtypes (Koonin, Makarova, & Zhang, 2017). The class 2 CRISPR–Cas systems that use a single Cas protein to recognize and degrade target DNA or RNA have been harnessed for a vast range of genetic engineering applications in various organisms. The DNA targeting CRISPR–Cas effector complexes locate their target site by scanning and interrogating the genomic DNA. The effector complexes are directed to specific DNA sequences by CRISPR RNA (crRNA) spacers that pair with the complementary segments in foreign DNA (protospacers), displacing the noncomplementary strand to form an R-loop (Jore et al., 2011; Szczelkun et al., 2014). Recognition of target DNA by CRISPR–Cas effector complexes requires a short protospacer adjacent motif (PAM) located near the targeted sequence (Jinek et al., 2012; Mojica, Diez-Villasenor, Garcia-Martinez, & Almendros, 2009). The type IIA Cas9 nuclease from Streptococcus pyogenes (SpCas9) is most widely used for genome editing. A Cas9 derivative that lacks endonuclease activity but can bind to targets (dCas9) is used for transcriptome modulation, base-specific genome editing and visualization of genomic loci in live cells (Chen et al., 2013; Gaudelli et al., 2017; Gilbert et al., 2013). A native dual-guide RNA (dgRNA) of Cas9 composed of the crRNA containing spacer sequence and partially complementary to crRNA trans-activating tracrRNA can be replaced with a single guide RNA (sgRNA) made up of fused crRNA and tracrRNA molecules (Jinek et al., 2012). The crystal structure of SpCas9 in complex with sgRNA and target DNA shows that the sgRNA assumes a T-shaped architecture comprising a spacer segment, a guide: target heteroduplex, a repeat: anti-repeat duplex and 3′-terminal stem loops (also known as bulge, nexus, and hairpins 1 and 2) (Briner et al., 2014; Nishimasu et al., 2014). Mechanisms of DNA interrogation and target cleavage by CRISPR–Cas systems have been extensively investigated. Studies of dynamics of SpCas9 DNA interrogation show that 3D diffusion dominates Cas9 genome searching and that off-target binding events are, on average, short-lived (Knight et al., 2015; Ma et al., 2016; Singh, Sternberg, Fei, Doudna, & Ha, 2016; Sternberg, Redding, Jinek, Greene, & Doudna, 2014). Biochemical and single-molecule experiments indicate that DNA strand separation and RNA/DNA heteroduplex formation by SpCas9 initiate at the protospacer end proximal to the 5′-NGG PAM sequence and proceed toward the distal end (Sternberg et al., 2014). The Cas9/PAM interaction plays a critical role during early stage of DNA interrogation by facilitating protospacer DNA strand separation near PAM and promoting initial base pairing between the target DNA strand and the RNA guide sequence (Mekler, Minakhin, & Severinov, 2017; Sternberg et al., 2014).
In addition to SpCas9, type V Cas12a (also known as Cpf1) and type IIC Cas9 nucleases from various bacteria can be advantageous in many applications (Fagerlund, Staals, & Fineran, 2015; Murugan, Babu, Sundaresan, Rajan, & Sashital, 2018; Ran et al., 2015; Zetsche et al., 2015). The molecular details of DNA interrogation and formation of the R-loop complex by CRISPR–Cas effectors are not fully understood. One way to address this problem is to investigate the effector interactions with target DNAs and model DNA probes in particular with probes that likely mimic interactions in intermediate effector-DNA complexes. Similar methodology has been applied to mechanisms of duplex DNA destabilization by a variety of enzymes that use base flipping to gain access to bases in the double-stranded DNA (Cao, Jiang, Stivers, & Song, 2004; Klimasauskas & Roberts, 1995). Measurement of CRISPR–Cas effector interactions with DNA probes by standard EMSA-based methods is hampered when specific DNA-effector affinity is low. To overcome this limitation, we developed a fluorometric assay based on measuring emission from fluorescently labeled derivatives of target DNA, which allows quantitative determination of both low and high specific affinity of the effector complexes for various DNA probes (Kuznedelov et al., 2016; Mekler et al., 2017). The assay can also be used to monitor the formation of CRISPR–Cas effector complexes (Mekler, Minakhin, Semenova, Kuznedelov, & Severinov, 2016). This chapter will describe the principle of the Cas beacon assay and several examples of its applications with emphasis on the methodical aspects.
2. Design and validation of Cas beacon method
2.1. Principle of the assay
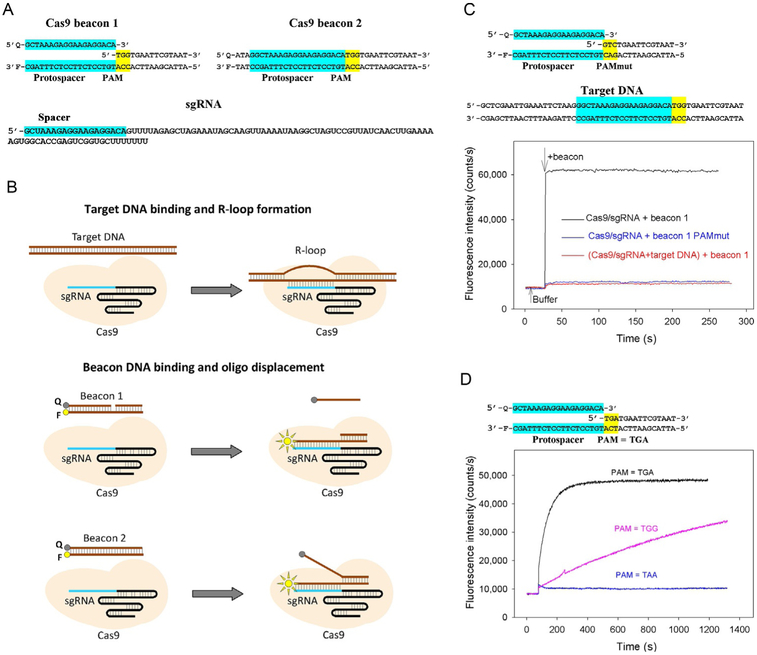
The fluorescently labeled target DNA derivatives to be used in the assays are prepared by annealing two or three oligonucleotides to generate a ~40-nt fragment of DNA comprising a protospacer, a functional PAM and ~15-bp DNA segment adjacent to PAM (Fig. 1A). We refer to these DNA constructs as “Cas beacons.” Beacon 1 shown in Fig. 1A consists of three oligonucleotides: its nontarget strand contains a discontinuity between PAM and the protospacer, while beacon 2 consists of two fully complementary oligonucleotides. The PAM-distal ends of the beacon target and nontarget strands are labeled with a fluorescent label and fluorescence quencher, respectively. The baseline fluorescence intensity of beacons is low because of proximity of the fluorescence label with the quencher. The CRISPR–Cas effector complexes bind to beacons in a way that mimics their binding to target DNA (Fig. 1B), separating the fluorophore and the quencher, which leads to a readily detectable increase in fluorescence intensity. The rate of binding and maximum increase in fluorescence intensity depend on the beacon sequence and structure.
Fig. 1.
A Cas beacon assay to detect Cas9/guide RNA complexes. (A) Structures of sgRNA and Cas9 beacon constructs. The PAM and protospacer sequences are highlighted in yellow and blue. (B) Schematic representation of target DNA and Cas9 beacons binding. The circles labeled F and Q indicate the fluorophore and quencher. (C) Measuring of Cas9/sgRNA complex binding to beacon 1. Time dependence of the increase in fluorescence upon the addition of 1nM beacon 1 or a control beacon 1 derivative bearing mutated PAM sequence to 3nM Cas9 preincubated with 5nM sgRNA for 30min. (D) Time dependence of the increase in fluorescence upon the addition of 1nM beacon 1 and its derivatives bearing TGA and TAA PAM sequences to a preincubated with sgRNA Cas9 derivative that recognizes TGA PAM sequence.
2.2. Validation of the Cas beacon assay
Initially, the beacon assay was developed for SpCas9 (Mekler et al., 2016). As seen in Fig. 1C, upon the addition of beacon 1 composed of three oligonucleotides to SpCas9 preincubated with sgRNA, fluorescence intensity increased by ~50-fold reaching peak intensity during the mixing time. Control experiments are needed to verify that the increase in fluorescence intensity is due to specific interaction between the effector and the beacon. In the case of Cas9/sgRNA, one can show that there is no change in fluorescence intensity upon the addition of beacon to sgRNA without Cas9. Second, a double-stranded DNA fragment containing SpCas9 PAM (5′-NGG) and a protospacer targeted by sgRNA spacer segment efficiently competes with the beacon for the binding to the Cas9/sgRNA complex (Fig. 1C), consistent with reported data indicating that Cas9 associates tightly with target DNA (Sternberg et al., 2014). Finally, fluorescence signal of beacon with a PAM mutation known to severely hinder the interaction of target DNA with SpCas9 does not increase in the presence of SpCas9/sgRNA complex (Fig. 1C).
Several SpCas9 variants engineered by molecular evolution exhibit altered PAM specificities (Kleinstiver et al., 2015). We monitored binding of an engineered SpCas9 variant that recognizes NGA PAM sequence to beacon 1 bearing TGG PAM and its derivatives that bore TGA and TAA PAMs. As expected, we found that the beacon bearing TGA PAM bound to the engineered SpCas9 much faster than beacon 1, while no signal increase was observed with beacon bearing TAA PAM sequence (Fig. 1D). Overall, these results show that the SpCas9 effector complex binding to the beacon truly mimics its specific target-binding activity. Almost identical kinetic traces were obtained when dSpCas9 was used instead of SpCas9 in the experiments with beacon 1 indicating that the wide separation of fluorophore-labeled and quencher-labeled ends of the DNA strands took place upon the beacon binding step. The kinetics of dSpCas9/sgRNA binding to beacon 2 composed of two oligonucleotides was similar to that observed with beacon 1 (Mekler et al., 2016).
2.3. Cas beacons for CRISPR–Cas systems other than SpCas9
In addition to the SpCas9 PAM sequence, beacons shown in Fig. 1A also serendipitously bear the PAM sequence of the Cas9 homolog from Streptococcus thermophilus (Sth) 5′-TGGTG (Gasiunas, Barrangou, Horvath, & Siksnys, 2012) (conserved nucleotides are underlined). Given that, we carried out beacon assay measurements with SthCas9 programmed with Sth guide RNA that had the same spacer sequence as sgRNA shown in Fig. 1A. The kinetics traces observed upon the addition of beacon 1 to the SthCas9/sgRNA complex were quite similar to those obtained with SpCas9. We also observed that beacons composed of three oligonucleotides (similar to beacon 1 in Fig. 1B) generated high fluorescence signals upon binding to Cas12a, Cas12b, and Escherichia coli Cascade effector complexes. However, binding of dCas12b and Cascade effectors to beacons composed of two oligonucleotides led only to modest ~twofold increases in fluorescence intensity, which may be attributable to a relatively short separation of their PAM-distal ends upon the beacon binding which makes the distance between fluorophore and quencher comparable with the Forster radius. Given these observations, it is preferable to use Cas beacons composed of three oligonucleotides in experiments with Cas12b and Cascade (Jain et al., in press; Kuznedelov et al., 2016) and possible other effectors.
2.4. Experimental procedures
2.4.1. Preparation of CRISPR–Cas proteins, guide RNAs, and DNA constructs
All proteins and guide RNAs were produced according to previously published methods. SpCas9 wild-type (wt) and dCas9 proteins were expressed in Escherichia coli BL21(DE3) using, respectively, expression plasmids pMJ806 and pMJ841 (Addgene, https://www.addgene.org/), and purified essentially as previously described (Jinek et al., 2012; Mekler et al., 2016). The expression plasmid encoding altered PAM specificity SpCas9 VQR variant was obtained by replacing wt cas9 gene with the mutant gene from plasmid MSP469 (Kleinstiver et al., 2015). Lachnospiraceae bacterium Cas12a (LbCas12a) was expressed in E. coli BL21(DE3) using expression plasmid pMAL-his-LbCpf1-EC (Addgene, Plasmid #79008) and purified essentially as previously described (Swarts, van der Oost, & Jinek, 2017).
Cas9 sgRNAs, tracrRNA, and crRNAs are made in vitro by T7 RNA polymerase (New England Biolabs) transcription from polymerase chain reaction-generated dsDNA templates according to manufacturer recommendations. Transcription products are purified using 10% denaturing polyacrylamide gel electrophoresis. Dual guide RNA was formed by mixing equimolar amounts of purified crRNA and tracrRNA in a buffer containing 40mM Tris, pH 7.9, 100mM NaCl; heating for 30s at 90°C and slowly cooling the reactions to 20°C. We also tested dgRNAs composed from tracrRNAs and crRNAs synthesized by Integrated DNA Technologies (IDT) and Sigma-Aldrich. In our experiments, they were indistinguishable from those prepared with T7 RNA polymerase. Cas12a crRNA was synthesized by IDT. Yeast tRNA and total RNA from human lung tissue were purchased from Thermo Fisher Scientific.
DNA probes were prepared from unmodified and chromophore-labeled DNA oligonucleotides synthesized by IDT. Target double-stranded (ds) DNA, competitor DNA probes and Cas beacon DNA constructs are formed by mixing equimolar amounts of synthetic complementary strands (final concentrations within low μM range) in a buffer containing 40mM Tris, pH 7.9, 100mM NaCl; heating for 1min at 90°C and slowly cooling the reactions in a real-time PCR machine with a temperature gradient (90–20°C, 1°C/min). The PAM-distal ends of the beacon target and nontarget strands are labeled with fluorescein and Iowa BlackFQ, respectively, introduced during commercial synthesis.
2.4.2. Fluorometric measurements
Fluorescence measurements are carried out at 25°C using a QuantaMaster QM4 spectrofluorometer (PTI) in binding buffer [20mM Tris HCl (pH 7.9), 100mM NaCl, 5% glycerol, 0.1mM DTT and 1mM MgCl2] containing 0.02% Tween-20. The Mg2+ concentration used was chosen since it is close to estimates of intracellular concentration of free Mg2+ (Rink,Tsien, & Pozzan, 1982). Final assay mixtures (800μL) contain 0.5–1nM Cas beacon, and Cas effector and guide RNAs at various concentrations. The fluorescein fluorescence intensities were recorded with an excitation wavelength of 498nm and an emission wavelength of 520nm. Time-dependent fluorescence changes were monitored using manual mixing; the mixing dead-time was ~15s. Competition experiments were analyzed with Felix software (PTI). The initial rates of beacon binding in the absence or presence of competitors were determined from the initial stage of the beacon-binding kinetic traces. The rate values were calculated as slopes of the kinetic curve segments from the mixing point to the time points at which beacon binding was about 15% complete. The Kd values are determined as averages obtained from at least three independent experiments.
3. Studies of assembly of CRISPR–Cas effector complexes
3.1. Using the Cas9 beacon to monitor Cas9/sgRNA complex assembly
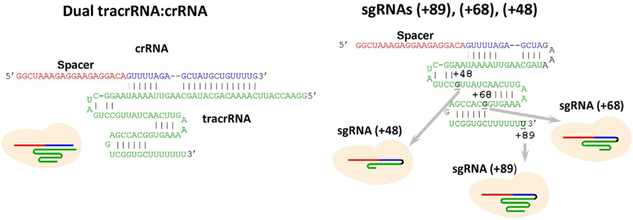
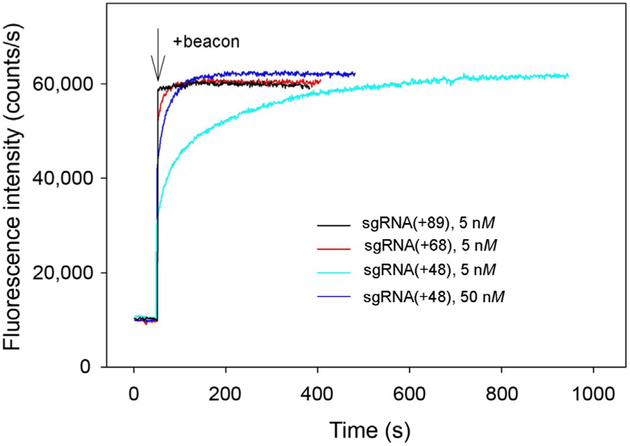
The delivery of Cas9/guide RNA system can be achieved by introducing vectors encoding nuclease and designed gRNA sequence into cell nuclei followed by Cas9 and gRNA production and the assembly of Cas9/gRNA complex. The assembly of gRNA into Cas9 may limit the efficiency of Cas9-mediated gene targeting, particularly in the case of less abundant gRNAs (Moreno-Mateos et al., 2015; Wang, Wei, Sabatini, & Lander, 2014). Kinetic and mechanistic characterization of gRNA loading into Cas9 in vitro may help to better understand mechanisms of the in vivo assembly process. To elucidate factors determining efficiency of the Cas9/guide RNA assembly process and in particular the role of 3′-terminal segment of guide RNA, we investigated the kinetics of Cas9 assembly with a dual guide RNA and several structurally distinct sgRNA versions (Fig. 2) and correlated the data with reported efficiencies of in vivo gene targeting by these guide RNAs. The sgRNA(+89) molecule (Fig. 2) is structurally similar to sgRNAs most widely used for CRISPR–Cas9-mediated muta-genesis and contains stem loops 1–3 of tracrRNA 3′ tail (nucleotides 23–89 of the native tracrRNA sequence), while sgRNA(+68) and sgRNA(+48) lack stem loop 3 or stem loops 2 and 3, respectively. The Cas9 beacon 1 shown in Fig. 1A was used to analyze target binding by effector complexes assembled with these guide RNAs. The kinetic traces for changes in fluorescence upon the binding of beacon to Cas9/sgRNA complexes obtained by incubation of 3nM Cas9 with 5nM of sgRNAs for 30min are shown in Fig. 3. The beacon rapidly bound to Cas9 complexes with sgRNA(+89) and sgRNA(+68). The kinetics of beacon binding to Cas9 preincubated with sgRNA(+48) was biphasic with distinct fast and slow components (Fig. 3). Increasing sgRNA(+48) concentration to 50nM decreased the amount of binding attributable to the slow component (Fig. 3). The kinetic behavior of Cas/sgRNA(+48) can be explained by lower affinity of sgRNA(+48) for Cas9 compared to other sgRNAs used (Wright et al., 2015).
Fig. 2.
Structures of guide RNAs.
Fig. 3.
Measuring of Cas beacon binding to Cas9 complexes with sgRNAs bearing 3′-tails of different lengths. Time dependence of the increase in fluorescence upon the addition of 1nM beacon to 3nM Cas9 preincubated with 5nM of sgRNA(+89), sgRNA(+68) and 5 or 50nM sgRNA(+48) for 30min.
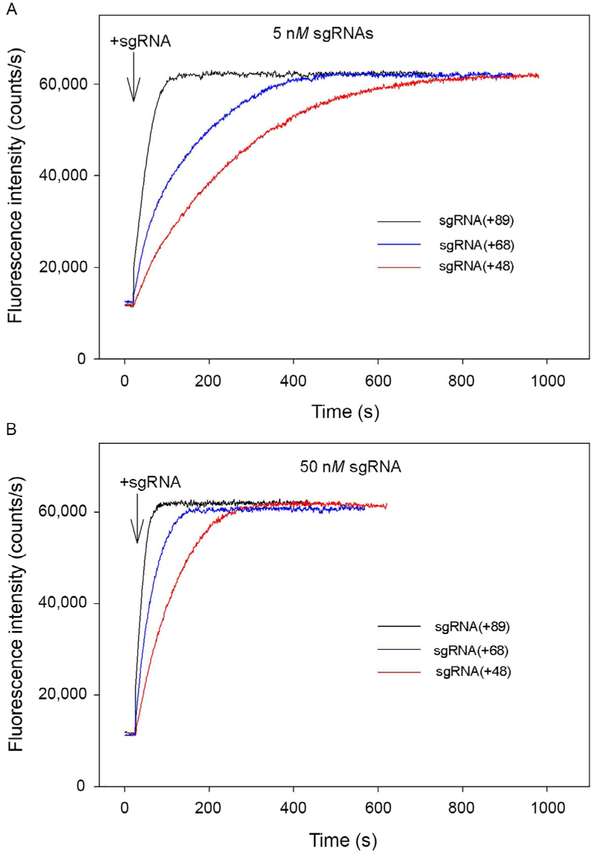
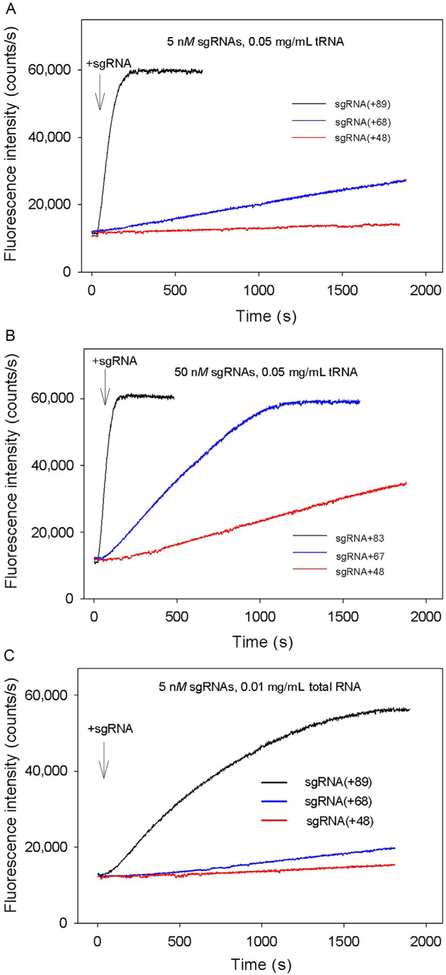
In experiments shown in Fig. 4, preincubation of Cas9 and sgRNA was omitted, and the kinetics of beacon binding was measured immediately upon the addition of sgRNAs to samples containing Cas9 and the beacon. The kinetic traces obtained in this way are considerably slower than corresponding traces measured with preformed Cas9/sgRNA complexes. The difference can be attributed to limitation of the rate of beacon binding by the relatively slow assembly of the Cas9/sgRNA complexes. In other words, traces in Fig. 4A and B closely represent the kinetics of formation of Cas9/sgRNA complexes capable of target recognition. As seen from data in Fig. 4, Cas9 most rapidly forms complexes with sgRNA(+89), whereas complexes with sgRNA(+68) and sgRNA(+48) form ~5- and 10-fold slower, respectively. Thus, the 3′-terminal stem loops 2 and 3 of sgRNA significantly affect the rate of sgRNA binding to Cas9.
Fig. 4.
Cas beacon assay for the assembly of sgRNAs into Cas9. Time dependence of the increase in fluorescence upon the addition of 5nM (A) or 50nM (B) sgRNAs to samples containing Cas9 and beacon.
Most proteins binding nucleic acids are capable of both specific and nonspecific binding. We examined whether formation of Cas9/sgRNA complexes can be affected by nonspecific.
RNA competitors using yeast tRNA and total RNA from human lung tissue. Most cellular RNA, with a notable exception of tRNA, is recruited into nucleoprotein complexes that are not expected to efficiently compete with sgRNA for the binding to Cas9. While intracellular concentrations of unbound RNAs are unknown, a pool of free RNA molecules must exist due to degradation of nucleoprotein complexes and other processes. With these considerations in mind, we used total human lung RNA at a concentration (0.01mg/mL) which is much lower than the typical total cellular RNA concentration (~10mg/mL; Priami & Morine, 2015). The concentrations of yeast tRNA were 0.05mg/mL. Incubation of preformed Cas9/sgRNA (+89) and Cas9/sgRNA(+68) complexes with RNA competitors for 2h had no effect on beacon binding (Fig. 5A). In contrast, incubation of preformed Cas9/sgRNA(+48) with nonspecific RNA caused a two- to threefold drop in beacon signal intensity (Fig. 5B). We conclude that sgRNA (+89) and sgRNA(+68) remain tightly bound to Cas9 once the complex is formed, whereas the Cas9/sgRNA(+48) complex is unstable in the presence of nonspecific RNA. When added to Cas9 before the addition of sgRNAs (5 or 50nM), the RNA competitors significantly delayed beacon binding (compare data in Figs. 6A–C and 4A and B). These data indicate that nonspecific RNAs readily bind to Cas9 and affect the Cas9/sgRNA assembly step. Total RNA produced larger effects than tRNA in these experiments (Fig. 6A and C), possibly because longer or differently structured nonspecific RNAs have a higher affinity for Cas9. Data in Fig. 6A–C show that nonspecific RNA affects the formation of Cas9 complexes with sgRNA(+68) and, most prominently, with sgRNA(+48) much stronger than with sgRNA(+89). Thus, low cleavage efficiency observed with sgRNA(+48) in eukaryotic cells may be at least in part a consequence of its inability to compete with intracellular RNA for binding to Cas9 (Hsu et al., 2013).
Fig. 5.
Effect of incubation of preformed Cas9/sgRNA complexes with nonspecific RNA competitors on beacon binding. Time dependencies of the increase in fluorescence upon the addition of 1nM beacon 1 to preformed Cas9/sgRNA(+89) and Cas9/sgRNA (+68) (A) or Cas9/sgRNA(+48) (B) complexes incubated for 2h with or without of nonspecific RNA competitors (0.05mg/mL yeast tRNA or 0.01mg/mL total human lung RNA) before the addition of beacon 1.
Fig. 6.
Effect of nonspecific RNA competitors on formation of Cas9/sgRNA complexes. (A and B) Time dependencies of the increase in fluorescence upon the addition of 5nM(A) or 50nM (B) sgRNAs to samples containing Cas9, beacon, and 0.05mg/mL yeast tRNA; (C) Time dependence of the increase in fluorescence upon the addition of 5nM sgRNAs to samples containing Cas9, beacon, and 0.01mg/mL total RNA from human lung.
3.2. Effect of salt on rate of the Cas9/sgRNA complex formation
Protein–nucleic acid interactions are sensitive to the concentrations and nature of electrolyte ions in solution (Leirmo, Harrison, Cayley, Burgess, & Record, 1987). We evaluated the influence of ion composition on beacon binding to preformed Cas9/sgRNA(+89) complex and on the Cas9/sgRNA(+89) assembly in assay mixtures containing 120mM of either NaCl, KCl, or KGlu. The kinetics of beacon bindings to preformed Cas9/sgRNA(+89) complex was rapid and similar to that shown in Fig. 4A at all conditions tested (data not shown). The effect of salt composition on kinetics of the Cas9/sgRNA(+89) assembly was measured in the presence of total human RNA similarly to experiments shown in Fig. 6C. The assembly half-times in samples containing NaCl, KCl, or KGlu were about 10, 20, and 40min, respectively (Fig. 7). Since mammalian cytosol typically contains higher concentration of K+ than Na+ and chloride is present only at low concentration (Lodish et al., 2000), the KGlu containing assay mixture may more closely mimic the intracellular ionic conditions.
Fig. 7.
Effect of salt composition on the Cas9 assembly with sgRNA(+89) in the presence of 0.01mg/mL total human lung RNA. The measurements were carried out in the assay mixtures containing 120mM of either NaCl, KCl, or KGlu.
3.3. Cas9 interactions with dgRNA
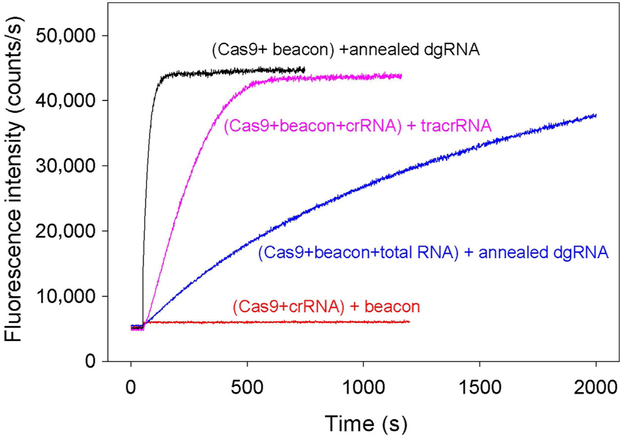
The dgRNA assembly into Cas9 was monitored as in experiments with sgRNAs by measuring beacon binding upon the addition of 5nM annealed crRNA and tracrRNA to samples containing Cas9 and beacon. As seen in Fig. 8, the beacon binding was fast, indicating rapid Cas9/dgRNA complex formation. In the presence of 0.01mg/mL total RNA, the kinetics of dgRNA assembly with Cas9 was delayed (t0.5 values are 25s and 14min, respectively) (Fig. 8). The relatively moderate effect of total RNA on the Cas9/dgRNA assembly as compared with that exerted on the Cas9/sgRNA(+48) complex formation (Fig. 6C) is consistent with a suggestion that stem loops 2 and 3 determine the ability of guide RNAs to withstand competition from nonspecific RNA. In agreement with reported inability of SpCas9 crRNA to function without tracrRNA (Deltcheva et al., 2011), no increase in fluorescence intensity was observed upon addition of the beacon to Cas9 preincubated with 50nM crRNA alone (Fig. 8). We also performed an order of addition Cas9/dgRNA assembly experiment in which beacon binding was measured upon the addition of tracrRNA to sample containing Cas9, beacon, and crRNA (Fig. 8). The kinetic trace observed in this experiment was ninefold slower (t0.5=220s) than the trace obtained with preannealed dgRNA, suggesting that the effector complex formation was limited by the crRNA/tracrRNA annealing step.
Fig. 8.
Cas9 assembly with dgRNA. Time dependencies of the increase in fluorescence upon the addition of annealed crRNA and tracrRNA to samples containing Cas9 and beacon in the absence or presence of 0.01mg/mL total human lung RNA (black and blue curves, respectively). Pink curve shows time dependence of the increase in fluorescence upon the addition of tracrRNA to sample containing Cas9, beacon, and crRNA. The crRNA and tracrRNA concentrations were 5nM in these experiments. No increase in fluorescence intensity was observed upon beacon addition to Cas9 preincubated with 50nM crRNA alone (red curve).
3.4. Using the Cas beacon assay to monitor Cas12a/crRNA complex assembly
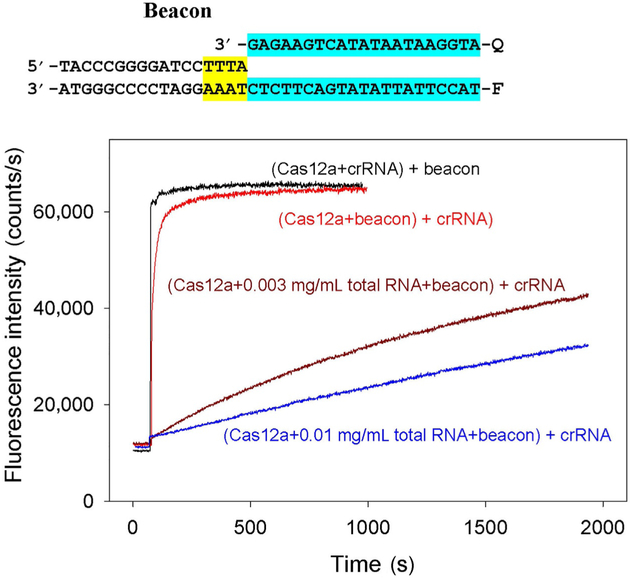
To determine how general is the sensitivity of CRISPR–Cas effector complex assembly to the presence of nonspecific RNA, effect of nonspecific RNA on the assembly of Cas12a with crRNA was investigated. The Cas12a effector complex requires a relatively short crRNA and does not need tracrRNA (Zetsche et al., 2015). The crRNA assembly with Cas12a was monitored in samples containing 100mM KGlu by measuring kinetics of beacon binding upon addition of crRNA to samples containing Cas12a and beacon. As in the case of Cas9/sgRNA assembly, the assay takes advantage of the observation that beacon binding to preformed Cas12a/crRNA complex is fast compared with rate of the Cas12a/crRNA complex formation. As shown in Fig. 9, formation of the Cas12a/crRNA complex was found to be considerably delayed by nonspecific RNA. The assembly half-times were about 0.3 and 50min, respectively, in the absence or presence of 0.01mg/mL total human lung RNA.
Fig. 9.
Effect of nonspecific RNA competitor on formation of Cas12a/crRNA complex. Time dependence of the increase in fluorescence upon the addition of 10nM crRNA to samples containing 5nM Cas12a, beacon, and 0.003 or 0.01mg/mL total RNA from human lung. The measurements were carried out in the assay mixtures containing 100mM of KGlu. Cas12a beacon structure is shown above the panel, the PAM and protospacer sequences are highlighted in yellow and blue.
Overall, the above data suggest that the rates of formation of Cas9/sgRNA and Cas12a/crRNA effector complexes in cells may be determined by competition between gRNA and free cellular RNAs for binding to the Cas9 and Cas12a enzymes. The loading of guide RNAs into Cas9 and Cas12a in KGlu containing buffer occurred on a ~1h time scale in the presence of 0.01mg/mL total human lung RNA (Figs. 7 and 9). Considering that concentrations of nonspecific RNA competitors in cells may be considerably higher than that used in this work, the effector complex assembly in cells may be even slower than it was observed in our experiments. Taking into account that unbound guide RNAs can be very unstable in eukaryotic cells (Ma et al., 2016) and that stability of both sgRNA and Cas9 in cells increases upon the complex formation (Harrington et al., 2017; Jinek et al., 2013; Moreno-Mateos et al., 2015), slow assembly may be a cause of significant loss of effector complexes due to degradation before the sgRNA loading takes place. This consideration is consistent with a report on high efficiency and accelerated rate of genome editing upon delivery of preassembled Cas9/sgRNA complexes in cells (Kim, Kim, Cho, Kim, & Kim, 2014).
4. Determination of CRISPR–Cas effectors affinities for DNA probes
Affinities of CRISPR–Cas/guide RNA complexes for target DNA and various model DNA substrates can be quantitatively determined by measuring the ability of these substrates to competitively affect the rate of CRISPR–Cas complexes binding to Cas beacons (Mekler et al., 2017). This approach was used to obtain quantitative information on target binding affinity of Cas12b effector complexes (Jain et al., in press) and E. coli Cascades assembled with different crRNAs (Kuznedelov et al., 2016). These studies demonstrated that the competition beacon assay allows one to quantitatively characterize low-affinity interactions even with intrinsically unstable effector complexes. Below we describe in detail the application of competition Cas beacon assay to study the mechanism of DNA interrogation by SpCas9 (Mekler et al., 2017).
4.1. DNA probes
To investigate the roles of distinct Cas9/sgRNA–DNA interactions during early stages of DNA interrogation, we determined binding affinities of dCas9/sgRNA to a set of double-stranded, partially single-stranded, or mismatched (bubbled) model substrates that mimic early Cas9/sgRNA–DNA intermediates on the pathway to the final R-loop complex (Mekler et al., 2017). The structures of competitor DNA probes and of the beacon are shown in Fig. 10. The parental “target probe” is a 47-bp DNA duplex containing a protospacer fully complementary to the sgRNA spacer sequence. Probe 1 in Fig. 10 is a derivative of the 47-bp duplex in which the protospacer segment sequence is substituted by a sequence noncomplementary to the sgRNA spacer. Other probes in Fig. 10 are fragments or derivatives of probe 1 that contain a PAM sequence but bear no sgRNA spacer sequence complementarity. The probes were preincubated with dCas9/sgRNA for 10min followed by the addition of beacon and measuring the kinetics of beacon binding to dCas9/sgRNA. Because measurement of relatively slow beacon-binding rates is more convenient, we used a beacon that had one mismatch with the sgRNA spacer sequence at the position immediately adjacent to PAM (Fig. 10). Binding of this beacon to dCas9 takes several minutes in the absence of competitors. All beacon-binding reactions in the presence of DNA competitors (probes) 1–12 (Fig. 10) were at least 90% complete in 5h, indicating that affinity to the beacon was much higher than to competitors and that beacon binding to Cas9/sgRNA was practically irreversible. In all experiments, concentrations of competitors were significantly higher than concentrations of either Cas9/sgRNA or beacon, which simplified the Kd calculation.
Fig. 10.
Structures of beacon and DNA probes used. The probe numbers used in the text are shown to the left of the sequences. The PAM sequences are highlighted in yellow.
4.2. Calculation of Kd
The Kd calculation is based on the assumption that the beacon binding rate in the presence of competitor DNA probe is proportional to concentration of effector complex molecules unoccupied by competitors. This implies that the rate of beacon binding in the absence of competitor (V0) is proportional to the Cas9/sgRNA concentration, whereas the binding rate in the presence of competitor (V1) is proportional to concentration of Cas9/sgRNA molecules that remain unbound to competitor; that is V0 = X × [Cas9/sgRNA] and V1 = X × ([Cas9/sgRNA] − [Cas9/sgRNA * DNA]), where Cas9/sgRNA * DNA is the Cas9/sgRNA complex with competitor and X is a proportionality coefficient. Consequently, the concentration of Cas9/sgRNA * DNA was calculated from Eq. (1):
| (1) |
The Kd for Cas9/sgRNA binding to DNA probes was calculated from the chemical equilibrium (Eq. 2):
| (2) |
The V0 and V1 rates were determined from the initial stage of the beacon-binding reactions. The rate values are calculated as slopes of the kinetic curve segments from the mixing point to the time points at which beacon binding was about 15% complete. The Kd calculation procedure assumes that the equilibrium binding between Cas9/sgRNA and the competitor is reached during the reaction incubation time. This assumption is validated by the observation that preincubation of Cas9/sgRNA with competitors before the addition of beacon for either 2 or 30min yielded identical beacon binding curves. The equilibrium balance between free and competitor-bound Cas9/sgRNA fractions may become disturbed at late stages of beacon binding if the Cas9/sgRNA complex with competitor dissociates slowly. Calculation of V0 and V1 from the initial stage of the beacon-binding reaction allows one to avoid this potential complication.
4.3. Interaction of dCas9/sgRNA with DNA probes without complementarity to sgRNA spacer
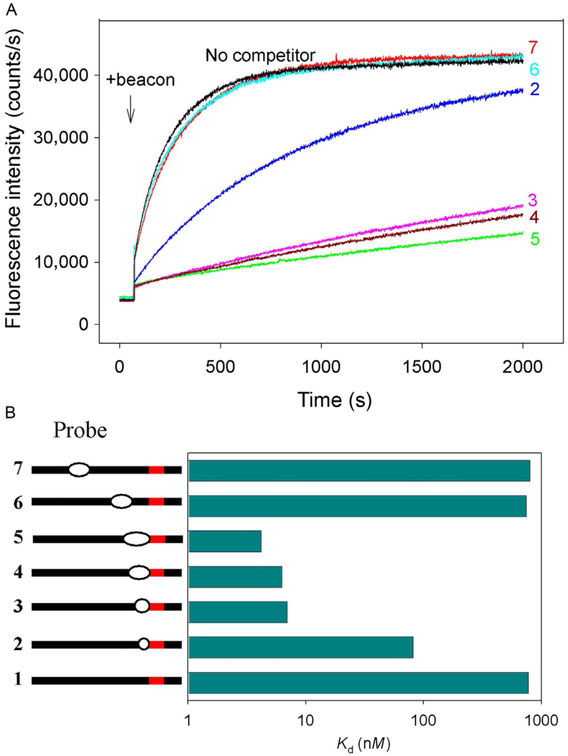
Unpairing of a short PAM-proximal segment of protospacer (1 or 2bp) is a critical step in DNA interrogation and may determine the overall rate of target location (Sternberg et al., 2014). DNA probes 2–7 carrying short mismatched “bubble” segments near PAM may mimic the DNA structure in early interrogation complexes and can be used as tool to study the target location/DNA interrogation mechanism. Assaying of dCas9/sgRNA interaction with probes 2–5 containing mismatched segments adjacent to PAM showed that 1- and 2-bp bubbles in probes 2 and 3, respectively, increase affinity about 10- and 100-fold compared with the parent probe (Fig. 11). Further extension of mismatched segments in probes 4 and 5 only slightly improves the affinity (about twofold). Mismatches in positions distant from PAM (probes 6 and 7) have no significant effect on the binding (Fig. 11). These data indicate that dCas9/sgRNA specifically binds with high affinity to double-stranded DNA bearing short unpaired segments adjacent to PAM. The increased affinity should cause a shift in the equilibrium between the duplex and unpaired conformations of the target and thus stimulate unpairing of a few base pairs nearest to PAM during the early stages of target interrogation even in the absence of pairing between the crRNA guide and the protospacer.
Fig. 11.
Competition assays with partially mismatched DNA probes. All probes bore no sgRNA guide sequence complementarity, and the concentrations of all probes were 200nM. (A) Representative data on effect of competitor DNA probes 2–7 that contained PAM mismatched “bubble” segments either adjacent to or distant from PAM on the kinetics of beacon binding to dCas9/sgRNA. (B) Quantification of competition data shown in (A).
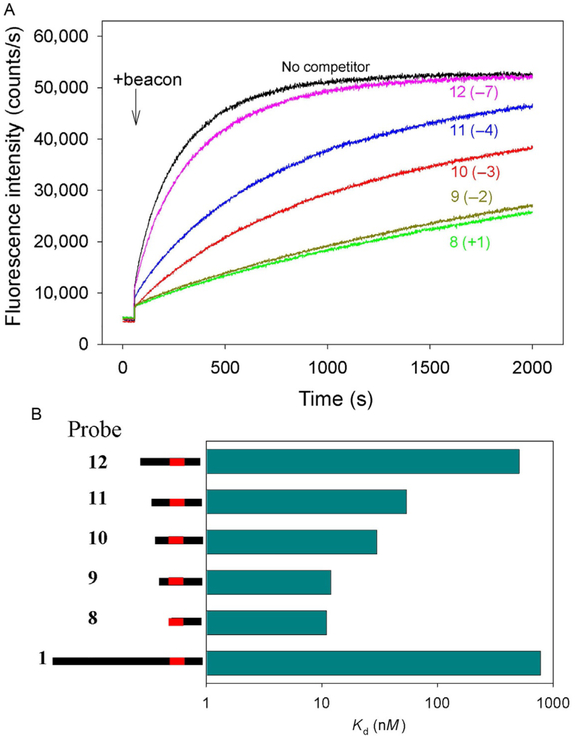
DNA bending has been proposed to facilitate DNA opening in various biological systems. In crystal structures of Cas9/sgRNA bound to target DNA, the DNA duplex containing the PAM sequence is bent toward the sgRNA spacer/DNA protospacer (Anders, Niewoehner, Duerst, & Jinek, 2014; Jiang et al., 2016). Further, domains 2 and 3 of the helical recognition lobe sterically occlude the central channel in the Cas9/sgRNA binary complex where the RNA/DNA hybrid is located in the ternary Cas9/sgRNA–DNA complexes (Jiang, Zhou, Ma, Gressel, & Doudna, 2015). This indicates that a structural rearrangement within the helical recognition lobe must occur upon target DNA binding and R-loop formation (Jiang et al., 2015). Thus, in early intermediate complexes duplex DNA may be sharply bent in the vicinity of PAM. To test this conjecture, we conducted competition binding experiments with duplex probes 8–12 that had the same downstream boundary but different upstream edges (Figs. 10 and 12). As shown in Fig. 12, DNA probe 8 bearing no nucleotides upstream of PAM and probe 9 with only a short 2bp duplex region upstream of PAM bind much stronger than the long probe 1. Extension of the upstream edge to positions −3 and −4 (probes 10 and 11) leads to gradual drops in the affinity, whereas probe 12 bearing a 7bp segment upstream of PAM is a weak competitor, similar to probe 1 (Fig. 12). The substitution of a G for a T at the +2 position of PAM in probe 8 virtually eliminates the competition effect, proving that probe 8 binding is PAM-dependent. These results indicate that intrinsic interaction of Cas9/sgRNA with PAM is quite strong. However, the presence of upstream DNA duplex noncomplementary to sgRNA guide beyond a certain point weakens this interaction. In other words, entry of dsDNA into the SpCas9/sgRNA complex is sterically hindered. This steric obstacle may promote target DNA bending and destabilization of the PAM-proximal duplex in the initial interrogation complex.
Fig. 12.
Competition assays using double-stranded DNA probes with upstream edges at different positions. (A) Effect of competitor DNA probes 8–12 on the kinetics of beacon binding to dCas9/sgRNA. Positions of upstream edges are shown in parentheses.(B) Quantification of competition data shown in (A). Bars are means, and errors are SDs (n = 3). All probes bore no sgRNA guide sequence complementarity. The concentrations of all competitor DNA probes were 200nM.
5. Conclusions
The examples presented above demonstrate how the simplest version of Cas beacon method allows one to study assembly of CRISPR–Cas surveillance complexes. To determine affinities of CRISPR–Cas complexes for various DNA probes, the basic assay shown in Fig. 1 needs to be modified by including competition between beacons and DNA probes of interest. This setup allows one to determine equilibrium dissociation constant values of effector complexes with target DNA and various DNA probes rapidly and precisely even if the affinity of the interaction is low. The advantages of the Cas beacon assay are that it reports only on functional effector complexes capable of specific binding to target and that the beacon binding produce large increases in fluorescence intensity (30- to 50-fold) which are considerably higher than fluorescence signals typically caused by nonspecific effects. The experiments presented above can be readily applied for analysis of assembly and target interrogation with any CRISPR–Cas effector and many additional experiments addressing important basic and practical questions can be performed with minimal modifications of either format of the assay.
Acknowledgments
This work was supported by grants from the National Institute of Health (GM10407 to K.S.), National Science Foundation (1652661 to D.G.S.), and National Institute of Food and Agriculture (IOW05480 to D.G.S.).
References
- Anders C, Niewoehner O, Duerst A, & Jinek M (2014). Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature, 513, 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, et al. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science, 315, 1709–1712. [DOI] [PubMed] [Google Scholar]
- Briner AE, Donohoue PD, Gomaa AA, Selle K, Slorach EM, Nye CH, et al. (2014). Guide RNA functional modules direct cas9 activity and orthogonality. Molecular Cell, 56, 333–339. [DOI] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, et al. (2008). Small CRISPR RNAs guide antiviral defense in prokaryotes. Science, 321, 960–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao C, Jiang YL, Stivers JT, & Song F (2004). Dynamic opening of DNA during the enzymatic search for a damaged base. Nature Structural and Molecular Biology, 11, 1230–1236. [DOI] [PubMed] [Google Scholar]
- Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, et al. (2013). Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell, 155, 1479–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, et al. (2011). CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature, 471, 602–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerlund RD, Staals RHJ, & Fineran PC (2015). The Cpf1 CRISPR-Cas protein expands genome-editing tools. Genome Biology, 16, 251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, et al. (2010). The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature, 468, 67–71. [DOI] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, & Siksnys V (2012). Cas9-crRNA ribonucleo-protein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proceedings of the National Academy of Sciences of the United States of America, 109, E2579–E2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, et al. (2017). Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature, 551, 464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, et al. (2013). CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell, 154, 442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington LB, Paez-Espino D, Staahl BT, Chen JS, Ma E, Kyrpides NC, et al. (2017). A thermostable Cas9 with increased lifetime in human plasma. Nature Communications, 8, 1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, et al. (2013). DNA targeting specificity of RNA-guided Cas9 nucleases. Nature Biotechnology, 31, 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain I, Minakhin L, Mekler V, Sitnik V, Rubanova N, Severinov K, et al. , Defining the seed sequence of the Cas12b CRISPR-Cas effector complex, RNA Biology, in press, 10.1080/15476286.2018.1495492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Taylor DW, Chen JS, Kornfeld JE, Zhou K, Thompson AJ, et al. (2016). Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science, 351, 867–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Zhou K, Ma L, Gressel S, & Doudna JA (2015). Structural biology. A Cas9-guide RNA complex preorganized for target DNA recognition. Science, 348, 1477–1481. [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, & Charpentier E (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science, 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, East A, Cheng A, Lin S, Ma E, & Doudna J (2013). RNA-programmed genome editing in human cells. eLife, 2, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, Waghmare SP, et al. (2011). Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nature Structural & Molecular Biology, 18, 529–536. [DOI] [PubMed] [Google Scholar]
- Kim S, Kim D, Cho SW, Kim J, & Kim JS (2014). Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Research, 24, 1012–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, et al. (2015). Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature, 523, 481–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimasauskas S, & Roberts RJ (1995). M. HhaI binds tightly to substrates containing mismatches at the target base. Nucleic Acids Research, 23, 1388–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SC, Xie L, Deng W, Guglielmi B, Witkowsky LB, Bosanac L, et al. (2015). Dynamics of CRISPR-Cas9 genome interrogation in living cells. Science, 350, 823–826. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS, & Zhang F (2017). Diversity, classification and evolution of CRISPR-Cas systems. Current Opinion in Microbiology, 37, 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznedelov K, Mekler V, Lemak S, Tokmina-Lukaszewska M, Datsenko KA, Jain I, et al. (2016). Altered stoichiometry Escherichia coli Cascade complexes with shortened CRISPR RNA spacers are capable of interference and primed adaptation. Nucleic Acids Research, 44, 10849–10861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leirmo S, Harrison C, Cayley DS, Burgess RR, & Record MT (1987). Replacement of potassium chloride by potassium glutamate dramatically enhances protein–DNA interactions in vitro. Biochemistry, 26, 2095–2101. [DOI] [PubMed] [Google Scholar]
- Lodish H, Berk A, Zipursky S, Matsudaira P, Baltimore D, & Darnell J (2000). Molecular cell biology. NY: WH. Freeman. [Google Scholar]
- Ma H, Tu LC, Naseri A, Huisman M, Zhang S, Grunwald D, et al. (2016). CRISPR-Cas9 nuclear dynamics and target recognition in living cells. Journal of Cell Biology, 214, 529–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekler V, Minakhin L, Semenova E, Kuznedelov K, & Severinov K (2016). Kinetics of the CRISPR-Cas9 effector complex assembly and the role of 3′-terminal segment of guide RNA. Nucleic Acids Research, 44, 2837–2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekler V, Minakhin L, & Severinov K (2017). Mechanism of duplex DNA destabilization by RNA-guided Cas9 nuclease during target interrogation. Proceedings of the National Academy of Sciences of the United States of America, 114, 5443–5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, & Almendros C (2009). Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology, 155, 733–740. [DOI] [PubMed] [Google Scholar]
- Moreno-Mateos MA, Vejnar CE, Beaudoin JD, Fernandez JP, Mis EK, Khokha MK, et al. (2015). CRISPR scan: Designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nature Methods, 12, 982–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugan K, Babu K, Sundaresan R, Rajan R, & Sashital DG (2018). The revolution continues: Newly discovered systems expand the CRISPR-Cas toolkit. Molecular Cell, 68, 15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, et al. (2014). Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell, 156, 935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priami C, & Morine MJ (2015). Analysis of biological systems. London: Imperial CollegePress. [Google Scholar]
- Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, et al. (2015). In vivo genome editing using Staphylococcus aureus Cas9. Nature, 520, 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink TJ, Tsien RY, & Pozzan T (1982). Cytoplasmic pH and free Mg2+ in lymphocytes. Journal of Cell Biology, 95, 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D, Sternberg SH, Fei J, Doudna JA, & Ha T (2016). Real-time observation of DNA recognition and rejection by the RNA-guided endonuclease Cas9. Nature Communications, 7, 12778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, Redding S, Jinek M, Greene EC, & Doudna JA (2014). DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature, 507, 62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts DC, van der Oost J, & Jinek M (2017). Structural basis for guide RNA processing and seed-dependent DNA targeting by CRISPR-Cas12a. Molecular Cell, 66, 221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczelkun MD, Tikhomirova MS, Sinkunas T, Gasiunas G, Karvelis T, Pschera P, et al. (2014). Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proceedings of the National Academy of Sciences of the United States of America, 111, 9798–9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, & Lander ES (2014). Genetic screens in human cells using the CRISPR-Cas9 system. Science, 343, 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AV, Sternberg SH, Taylor DW, Staahl BT, Bardales JA, Kornfeld JE, et al. (2015). Rational design of a split-Cas9 enzyme complex. Proceedings of the National Academy of Sciences of the United States of America, 112, 2984–2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, et al. (2015). Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell, 163, 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]