Abstract
Pseudouridine (Ψ) is the most abundant RNA modification and has been found in many kinds of RNAs, including snRNA, rRNA, tRNA, mRNA, and snoRNA. Thus, Ψ sites play a significant role in basic research and drug development. Although some experimental techniques have been developed to identify Ψ sites, they are expensive and time consuming, especially in the post-genomic era with the explosive growth of known RNA sequences. Thus, highly accurate computational methods are urgently required to quickly detect the Ψ sites on uncharacterized RNA sequences. Several predictors have been proposed using multifarious features, but their evaluated performances are still unsatisfactory. In this study, we first identified Ψ sites for H. sapiens, S. cerevisiae, and M. musculus using the sequence features from the bi-profile Bayes (BPB) method based on the random forest (RF) and support vector machine (SVM) algorithms, where the performances were evaluated using 5-fold cross-validation and independent tests. It was found that the SVM-based accuracies were 3.55% and 5.09% lower than the iPseU-CUU predictor for the H_990 and S_628 datasets, respectively. Almost the same-level results were obtained for M_994 and an independent H_200 dataset, even showing a 5.0% improvement for S_200. Then, three different kinds of features, including basic Kmer, general parallel correlation pseudo-dinucleotide composition (PC-PseDNC-General), and nucleotide chemical property (NCP) and nucleotide density (ND) from the iRNA-PseU method, were combined with BPB to show their comprehensive performances, where the effective features are selected by the max-relevance-max-distance (MRMD) method. The best evaluated accuracies of the combined features for the S_628 and M_994 datasets were achieved at 70.54% and 72.45%, which were 2.39% and 0.65% higher than iPseU-CUU. For the S_200 dataset, it was also improved 8% from 69% to 77%. However, there was no obvious improvement for H. sapiens, which was evaluated as approximately 63.23% and 72.0% for the H_990 and H_200 datasets, respectively. The overall performances for Ψ identification using BPB features as well as the combined features were not obviously improved. Although some kinds of feature extraction methods based on the RNA sequence information have been applied to construct the predictors in previous studies, the corresponding accuracies are generally in the range of 60%–70%. Thus, researchers need to reconsider whether there is any sequence feature in the RNA Ψ modification prediction problem.
Keywords: pseudouridine site, bi-profile Bayes, random forest, support vector machine, max-relevance-max-distance method
Introduction
Pseudouridine (Ψ) is the most prevalent post-transcriptional modification, and it has been widely found in a series of biological and cellular processes.1,2 Recent studies have demonstrated that Ψ sites exist in many kinds of RNAs, such as small nuclear RNA (snRNA), rRNA, tRNA, mRNA, and small nucleolar RNA (snoRNA).3, 4, 5, 6, 7, 8, 9, 10, 11 Thus, the Ψ site plays a crucial role in biological research and drug development. More specifically, Ψ is an isomer of uridine catalyzed by the Ψ synthase (PUS) that removes the uridine residue’s base from its sugar, followed by “rotating” it 180° along the N3-C6 axis, and subsequently reattaches the base’s 5-carbon to the 1’-carbon of the sugar.12
Although there are several experimental methods based on the high-throughput techniques that have been developed to recognize the Ψ modifications, they are both costly and time consuming.13, 14, 15, 16, 17 In addition, researchers are facing an explosive increase of RNA data in the post-genomic age.18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30 Therefore, intelligent computational approaches are highly desirable to predict Ψ sites on RNA sequences.
To the best of our knowledge, six predictors have been reported to identify Ψ sites. Specifically, Panwar and Raghava31 first proposed the tRNAmod model to predict Ψ sites in tRNA. Li et al.32 then developed the PPUS method based on the support vector machine (SVM) to identify PUS-specific Ψ sites. Later, Chen et al.33 provided the iRNA-PseU predictor, and He et al.34 introduced the PseUI predictor, which are both based on the SVM classifier. In addition, Tahir et al.35 built the iPseU-CUU model based on the convolution neural network (CNN). Most recently, Chen et al.36 proposed an eXtreme Gradient Boosting (xgboost)-based method (XG-PseU). It should be noted that the same datasets, built by Chen et al.,33 were applied in the three studies (iRNA-PseU, PseUI, and iPseU-CUU) to build the predictors, including the benchmark training datasets (H_990, S_628, and M_944) and the independent testing datasets (H_200 and S_200). Here, H, S, and M represent the RNA samples for H. sapiens, S. cerevisiae, and M. musculus, while 990, 628, 944, and 200 indicate the corresponding sample numbers in each dataset. Thus, we used the datasets mentioned earlier in this article for convenient comparisons. The performances of the four predictors (iRNA-PseU, PseUI, iPseU-CUU, and XG-PseU) are listed in Table 1, where the XG-PseU results for independent datasets were obtained by the web server at http://www.bioml.cn. The jackknife test, 5-fold cross-validation, and 10-fold cross-validation are used for the iRNA-PseU, PseUI/iPseU-CUU, and XG-PseU models, respectively. It can be seen that their overall performances are gradually improved through the scientists’ efforts. Taking H_990 as an example, the accuracies have been improved by 6.28% from 60.40% (iRNA-PseU) to 61.24% (PseUI) and to 66.68% (iPseU-CUU). However, it must be noted that these predictive accuracies are still unsatisfactory.
Table 1.
Results of the Proposed iRNA-PseU, PseUI, iPseU-CUU, and XG-PseU Predictors for Training Datasets H_990, S_628, and M_944 and Testing Datasets H_200 and S_200
| Predictors | Training Datasets | Acc (%) | MCC | Sn (%) | Sp (%) | Testing Datasets | Acc (%) | MCC | Sn (%) | Sp (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| iRNA-PseUa | H_990 | 60.4 | 0.21 | 61.01 | 59.8 | H_200 | 65.00 | 0.30 | 60.00 | 70.00 |
| PseUIb | 64.24 | 0.28 | 64.85 | 63.64 | 65.50 | 0.31 | 63.00 | 68.00 | ||
| iPseU-CUUc | 66.68 | 0.34 | 65.00 | 68.78 | 69.00 | 0.40 | 77.72 | 60.81 | ||
| XG-PseUd | 65.44 | 0.31 | 63.64 | 67.24 | 67.00 | 0.34 | 67.00 | 67.00 | ||
| iRNA-PseUa | S_628 | 64.49 | 0.29 | 64.65 | 64.33 | S_200 | 73.00 | 0.46 | 81.00 | 65.00 |
| PseUIb | 66.56 | 0.33 | 62.1 | 71.02 | 68.50 | 0.37 | 72.00 | 65.00 | ||
| iPseU-CUUc | 68.15 | 0.37 | 66.36 | 70.45 | 73.50 | 0.47 | 68.76 | 77.82 | ||
| XG-PseUd | 68.15 | 0.37 | 66.84 | 69.45 | 71.00 | 0.42 | 75.00 | 67.00 | ||
| iRNA-PseUa | M_944 | 69.07 | 0.38 | 73.31 | 64.83 | |||||
| PseUIb | 70.44 | 0.41 | 74.58 | 66.31 | ||||||
| iPseU-CUUc | 71.81 | 0.44 | 74.49 | 69.11 | ||||||
| XG-PseUd | 72.03 | 0.45 | 76.48 | 67.57 |
As a crucial step toward building a machine-learning-based predictor, feature extraction becomes a particularly important process. Several sequence representation methods have been used in previous works to obtain feature vectors. For example, a hybrid approach of the binary profile of patterns (BPP) and structural information is applied in the tRNAmod.31 In addition, the PPUS model uses the nucleotides around Ψ as the features to identify.32 For the successful iRNA-PseU method, dinucleotide chemical properties (DCP) and nucleotide density (ND) are incorporated for identification.33 For the PseUI, the effective features are selected from five different feature extraction techniques using the sequential forward-feature-selection method, including nucleotide composition (NC), dinucleotide composition (DNC), pseudo-dinucleotide composition (PseDNC), position-specific nucleotide propensity (PSNP), and position-specific dinucleotide propensity (PSDP).37,38 For the iPseU-CUU method, the features are obtained automatically by a CNN model based on a deep learning machine, which is widely used in bioinformatics.39, 40, 41, 42 Furthermore, two additional feature extraction techniques, n-gram and multivariate mutual information (MMI), are also applied for the machine learning approach by the SVM method, where they still give a low accuracy (Acc).35 For the newly reported XG-PseU predictor, six feature extraction techniques are used, namely, NC, DNC, trinucleotide composition (TNC), nucleotide chemical property (NCP), ND, and one-hot encode (one hot).
At the same time, the identification of many types of RNA modifications using the machine-learning-based computational approaches shows the excellent performance, including for N6-methyladenosine (m6A),43, 44, 45 5-methylcytosine (m5C),46, 47, 48, 49, 50, 51, 52, 53 N1-methyladenosine (m1A),54, 55, 56 and so forth. The related kinds of computational models used for these purposes have been summarized in a review,57 in which the recently reported overall accuracies are basically above 90%. In particular, the SVM-based iRNA(m6A)-PseDNC model demonstrates an Acc of 91.24% of 10-fold cross-validation for m6A identification for S. cerevisiae.43 For the m5C site, the recently developed iRNA-m5C predictor by the Random Forest (RF) algorithm shows a jackknife test Acc up to 92.9% for H. sapiens.52 For m1A, the SVM-based iRNA-3typeA method obtains a jackknife validation Acc of 99.13% on H. sapiens and 98.73% for M. musculus.56 However, as mentioned earlier, the evaluated accuracies of Ψ site identification of different models are basically only 60%–70%, where there is still a large amount of improvement possible.
We noticed that a predictor called “KELMPSP” reported a better performance, where the accuracies for the H_990, S_628, M_949, H_200, and S_200 datasets are up to 74.55%, 85.53%, 79.45%, 72.5%, and 76.00%, respectively.58 In this method, the kernel extreme learning machine (KELM) algorithm is applied, where the final features are obtained by combining NCP, nucleotide concentrations, and position-specific mononucleotide, dinucleotide, and trinucleotide propensity characteristics. However, the related web server at http://39.105.77.161:8890/KELMPSP is no longer available.
In this paper, we first applied the bi-profile Bayes method (BPB)59 to extract the RNA sequence features to identify the Ψ sites. Two algorithms, RF and SVM, were both used to construct the models, where the performances were evaluated by 5-fold cross-validation and independent tests. Then, we incorporated three different features with BPB to show their comprehensive performance, including basic Kmer (Kmer),60 general parallel correlation pseudo-dinucleotide composition (PC-PseDNC-General) generated from the web server Pse-in-One,61 and NCP with ND (NCP+ND). Also, high-quality features were selected using the MRMD62 method to predict the Ψ sites.
Results and Discussion
Performance of the BPB Features
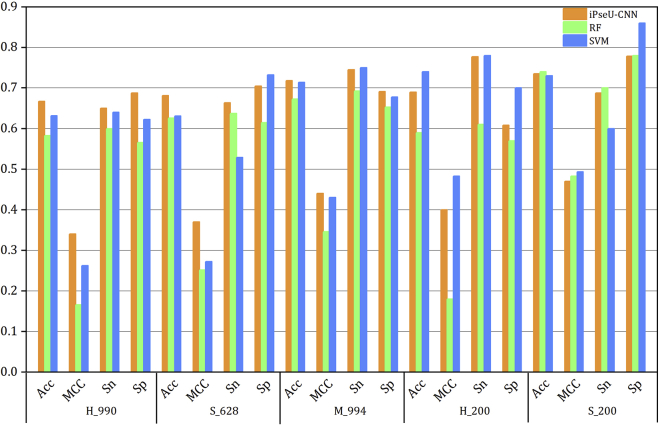
First, we extracted the RNA features using the BPB method for Ψ site prediction. The performances were evaluated over the 5-fold cross-validation for the benchmark datasets H_990, S_628, and M_944 and independent dataset for H_200 and S_200. Table 2 gives a comparison of our results using BPB features with the iPseU-CUU predictor, where RF and SVM indicate the results from the RF and SVM classifiers, respectively. It is obvious that the SVM generally performed better than the RF. Specifically, the accuracies of the SVM method were improved 4.85% and 4.13% for the training datasets H_990 and M_994, respectively. For the independent dataset test, the Acc and MCC were obviously increased by 15.0% and 0.3 for the H_200 datasets. However, for the S_628 datasets, the Acc was only increased from 62.58% to 63.06%. Here, we found that, although the specificity (Sp) increased from 61.46% to 73.25%, the sensitivity (Sn) actually declined from 63.69% to 52.87%, which means that one half of the positive samples were incorrectly predicted to be the false one. Similar results can also be observed in S_200. From the comparison between RF and SVM, it can be concluded that the SVM algorithm is more efficient than RF for the Ψ prediction of RNA sequences for H. sapiens and M. musculus.
Table 2.
Comparison of Our Results based on the RF and SVM Methods Using the BPB Features with the iPseU-CUU Predictor
| Predictors | Training Datasets | Acc (%) | MCC | Sn (%) | Sp (%) | Testing Datasets | Acc (%) | MCC | Sn (%) | Sp (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| iPseU-CNNa | H_990 | 66.68 | 0.34 | 65.00 | 68.78 | H_200 | 69.00 | 0.40 | 77.72 | 60.81 |
| RFb | 58.28 | 0.17 | 60.00 | 56.57 | 59.00 | 0.18 | 61.00 | 57.00 | ||
| SVMc | 63.13 | 0.26 | 64.04 | 62.22 | 74.00 | 0.48 | 78.00 | 70.00 | ||
| iPseU-CNNa | S_628 | 68.15 | 0.37 | 66.36 | 70.45 | S_200 | 73.50 | 0.47 | 68.76 | 77.82 |
| RFb | 62.58 | 0.25 | 63.69 | 61.46 | 74.00 | 0.48 | 70.00 | 78.00 | ||
| SVMc | 63.06 | 0.27 | 52.87 | 73.25 | 73.00 | 0.49 | 60.00 | 86.00 | ||
| iPseU-CNNa | M_944 | 71.81 | 0.44 | 74.49 | 69.11 | |||||
| RFb | 67.27 | 0.35 | 69.28 | 65.25 | ||||||
| SVMc | 71.40 | 0.43 | 75.00 | 67.80 |
The predictor proposed by Tahir et al.35
The RF-based predictor using BPB features.
The SVM-based predictor using BPB features.
Compared with the iPseU-CUU model, the SVM method showed accuracies reduced by 3.55% and 5.09% for the first two training datasets H_990 and S_628. Almost the same results could be found for the training dataset M_994 and independent dataset H_200, where our results are only 0.5% lower than that for iPseU-CUU. Additionally, the SVM model performed better for S_200, where the Acc and MCC were both improved approximately 5.0% and 0.08, respectively. In general, the SVM algorithm appears to be a better choice than RF for the Ψ modification prediction using BPB features alone, which can be clearly found in Figure 2. However, it must be noted that the overall performance of the SVM method here is unsatisfactory, even lower than that of the latest predictor, iPseU-CUU, for the two datasets H_990 and S_628.
Figure 2.
This Histogram Shows the Results of the iPseU-CUU Predictor and the Constructed Model Based on the RF and SVM Classifiers Using the BPB Features
Performance of the BPB Features Combining Other Features
For a better performance, three different kinds of features were also investigated: Kmer,60 PC-PseDNC-General, and NCP+ND from the iRNA-PseU method.33 At the same time, those features were further combined with BPB to achieve a better result, where the MRMD method was applied to select the important features for experiments.62 Table 3 lists the results of different feature selection for H_990 datasets using the RF method (left) and SVM method (right).
Table 3.
Results of Feature Selection for the H_990 Dataset Using the RF and SVM Methods
| Feature Subset | RF |
SVM |
||||||
|---|---|---|---|---|---|---|---|---|
| Acc (%) | MCC | Sn (%) | Sp (%) | Acc (%) | MCC | Sn (%) | Sp (%) | |
| BPB | 58.28 | 0.17 | 60.00 | 56.57 | 63.13 | 0.26 | 64.04 | 62.22 |
| Kmer(2) | 55.76 | 0.12 | 53.13 | 58.38 | 60.00 | 0.23 | 41.82 | 78.18 |
| Kmer(3) | 58.79 | 0.18 | 58.59 | 58.99 | 59.70 | 0.20 | 53.94 | 65.45 |
| Kmer(4) | 58.59 | 0.17 | 59.39 | 57.78 | 57.27 | 0.15 | 56.57 | 57.98 |
| PC-PseDNC-General (6,0.99) | 58.59 | 0.17 | 56.57 | 60.61 | 57.78 | 0.16 | 49.49 | 66.06 |
| NCP+ND | 56.87 | 0.14 | 57.37 | 56.36 | 60.34 | 0.21 | 60.40 | 60.28 |
| BPB+Kmer(3) | 60.40 | 0.21 | 60.61 | 60.20 | 63.23a | 0.27a | 61.01a | 65.45a |
| BPB+PC-PseDNC-General (6,0.99) | 61.72 | 0.23 | 59.39 | 64.04 | 62.93 | 0.26 | 61.62 | 64.24 |
| BPB+NCP+NP | 61.11 | 0.22 | 62.83 | 59.39 | 61.11 | 0.22 | 58.79 | 63.43 |
| BPB+PC-PseDNC-General (6,0.99) + Kmer(3) | 61.01 | 0.22 | 59.39 | 62.63 | 62.73 | 0.25 | 61.82 | 63.64 |
Performance with maximum accuracy.
The first six rows give the performance of each type of feature, including BPB, Kmer (k = 2, 3, 4), PC-PseDNC-General (λ = 6,w = 0.99), and NCP+ND. For the Kmer method,60 three results with k = 2, 3, and 4 are listed. It can be found that the Kmer(3) shows consistent results, where the accuracies are 58.79% and 59.70% for the RF and SVM classifiers, respectively. In the PC-PseDNC-General method,63,64 several parameters have been tested, and better results are obtained with the parameters λ = 6 and w = 0.99. The corresponding SVM-based Acc (57.78%) is slightly lower than the RF-based Acc (58.59%). We also repeated the work by Chen et al.54 (NCP+ND) with the 5-fold cross-validation, which obtained an Acc of 60.34% compared to the reported jackknife results (60.40%). From the discussion earlier, the performances of the single features are all lower than that of the latest iPseU-CUU predictor (66.68%), among which the BPB features give the best Acc (63.13%) by the SVM method.
Further, we combined the Kmer, PC-PseDNC-General, and NCP+ND features with the BPB, and the final useful features for model constructing were selected using the MRMD method.62 There were four results for the combined features listed in Table 3 for the H_990 datasets, including BPB+Kmer(3), BPB+PC-PseDNC-General(6,0.99), BPB+NCP+ND, and BPB+PC-PseDNC-General(6,0.99)+Kmer(3). It can be found that the combined results are generally improved 2%–3% over the single BPB results by the RF method, where the best combination with a maximum Acc 61.72% is BPB+PC-PseDNC-General(6,0.99). However, there is no obvious improvement for the SVM-based method and even a 2.02% decrease for the BPB+NCP+ND combination. The feature combination BPB+Kmer(3) showed the best performance by the SVM method, which gave 63.23% Acc, 0.27 MCC, 61.01% Sn, and 65.45% Sp. Applying this model to an independent test for H_200, the obtained Acc, MCC, Sp, and Sn were 72.00%, 0.46, 82%, and 62%, respectively. Compared to the iPseU-CUU predictor, 3% and 0.06 improvement for the Acc and MCC were found.
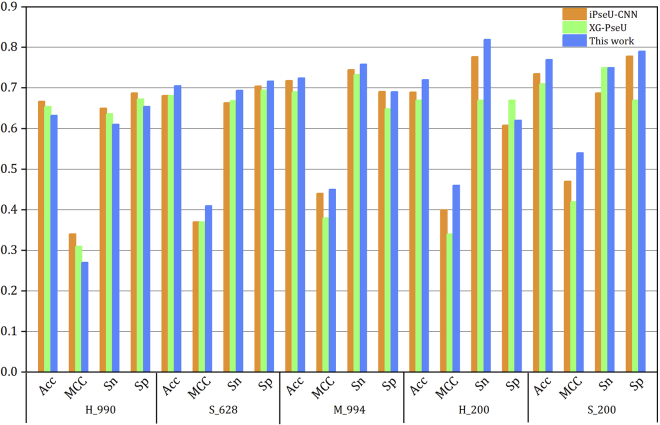
Tables 4 and 5 list the same results as in Table 3 but for the datasets S_628 and M_944, respectively. For S_628, the feature combination BPB+PC-PseDNC-General(2,0.1)+Kmer(4) gave the best performance, where the ACc and MCC were obviously improved by 7.48% and 0.14, respectively. When compared with the iPseU-CNN model, the evaluated Acc shows 2.39% improvement. Finally, the combined model was tested using the independent dataset S_200, where the Acc, MCC, Sn, and Sp are 77.00%, 0.54, 75%, and 79%, respectively. It can be seen that there were 3.5% and 0.07 improvement for the Acc and MCC compared to those for the iPseU-CUU model. For M_994, the best performance was given by feature combination BPB+Kmer(3), for which the Acc was 72.46%, MCC was 0.45, Sn was 75.85%, and Sp was 69.07%. Compared with the Acc of the iPseU-CUU method, there was only 0.65% improvement obtained. Figure 3 shows an intuitive comparison of the evaluated performance of the iPseU-CUU (orange bars), XG-PseU (green bars), and the constructed SVM-based model using the combined features in this work (blue bars).
Table 4.
Results of Feature Selection for the S_628 Dataset Using the RF and SVM Methods
| Feature Subset | RF |
SVM |
||||||
|---|---|---|---|---|---|---|---|---|
| Acc (%) | MCC | Sn (%) | Sp (%) | Acc (%) | MCC | Sn (%) | Sp (%) | |
| BPB | 62.58 | 0.25 | 63.69 | 61.46 | 63.06 | 0.27 | 52.87 | 73.25 |
| Kmer (k = 2) | 58.12 | 0.16 | 58.28 | 57.96 | 61.78 | 0.24 | 64.33 | 59.24 |
| Kmer (k = 3) | 60.35 | 0.21 | 62.10 | 58.60 | 61.78 | 0.24 | 66.56 | 57.01 |
| Kmer (k = 4) | 59.71 | 0.19 | 62.74 | 56.69 | 64.97 | 0.30 | 67.52 | 62.42 |
| PC-PseDNC-General (2, 0.11) | 58.76 | 0.18 | 61.78 | 55.73 | 61.15 | 0.22 | 64.01 | 58.28 |
| NCP+ND | 60.83 | 0.22 | 62.74 | 58.92 | 60.99 | 0.22 | 57.01 | 64.97 |
| BPB+Kmer (k = 4) | 64.01 | 0.28 | 64.33 | 63.69 | 68.15 | 0.36 | 66.56 | 69.75 |
| BPB+PC-PseDNC-General (2, 0.11) | 62.90 | 0.26 | 63.38 | 62.42 | 66.08 | 0.33 | 57.64 | 74.52 |
| BPB+NCP+ND | 62.74 | 0.26 | 65.61 | 59.87 | 61.78 | 0.24 | 56.37 | 67.20 |
| BPB+PC-PseDNC-General (2, 0.11) + Kmer(4) | 64.49 | 0.29 | 65.92 | 63.06 | 70.54a | 0.41a | 69.43a | 71.66a |
Performance with maximum accuracy.
Table 5.
Results of Feature Selection for the M_944 Dataset Using the RF and SVM Methods
| Feature Subset | RF |
SVM |
||||||
|---|---|---|---|---|---|---|---|---|
| Acc (%) | MCC | Sn (%) | Sp (%) | Acc (%) | MCC | Sn (%) | Sp (%) | |
| BPB | 68.54 | 0.37 | 69.28 | 67.80 | 71.40 | 0.43 | 75.00 | 67.80 |
| Kmer(2) | 52.22 | 0.04 | 54.45 | 50.00 | 56.78 | 0.14 | 61.65 | 51.91 |
| Kmer(3) | 55.51 | 0.11 | 57.42 | 53.60 | 59.22 | 0.18 | 60.81 | 57.63 |
| Kmer(4) | 56.04 | 0.12 | 58.05 | 54.03 | 58.37 | 0.17 | 59.96 | 56.78 |
| PC-PseDNC-General (2, 0.1) | 53.07 | 0.06 | 56.14 | 50.00 | 57.84 | 0.16 | 64.41 | 51.27 |
| NCP+ND | 67.58 | 0.35 | 70.34 | 64.83 | 68.01 | 0.36 | 69.49 | 66.53 |
| BPB+Kmer(3) | 67.37 | 0.35 | 71.61 | 63.14 | 72.46a | 0.45a | 75.85a | 69.07a |
| BPB+PC-PseDNC-General (2, 0.1) | 67.58 | 0.35 | 70.97 | 64.19 | 71.40 | 0.43 | 73.52 | 69.28 |
| BPB+NCP+ND | 68.43 | 0.37 | 71.82 | 65.04 | 68.11 | 0.36 | 69.70 | 66.53 |
| BPB+PC-PseDNC-General (2, 0.11) + Kmer(3) | 68.33 | 0.37 | 72.67 | 63.98 | 71.72 | 0.44 | 75.00 | 68.43 |
Performance with maximum accuracy.
Figure 3.
Comparisons of the Evaluated Performance of Predictors iPseU-CUU, XG-PseU and the Constructed Model Using the Combined Features in This Work
Finally, we investigated several kinds of features from the two state-of-the-art tools iLearn65 and BioSeq-Analysis2.066 for H. sapiens, including Mismatch (k = 2,3,4), subsequence (k = 2,3,4), the enhanced nucleic acid composition (ENAC) with the sequence window 5, electron-ion interaction pseudopotentials of trinucleotide (EIIP), electron-ion interaction pseudopotentials of trinucleotide (PseEIIP), binary encoding (BE), dinucleotide-based auto covariance (DAC), dinucleotide-based cross covariance (DCC), and dinucleotide-based auto-cross covariance (DACC). It was found that the average Acc of subsequence, ENAC, and autocorrelation features using the SVM algorithm is approximately 55%. The evaluated performances of other features as well as the combined features with the best performances BPB+Kmer(3) are listed in Table 6. It can be seen that the feature combination BPB+Kmer(3)+EIIP gives the accuracies 63.33% and 75% on the H_990 and H_200 datasets, which are improved by 0.1% and 3% compared with our original feature combination BPB+Kmer(3), respectively.
Table 6.
Results of Feature Selection for H_990 and H_200 Datasets Using Several Kinds of Features from iLearn and BioSeq-Analysis 2.0
| Feature Subset | H_990 |
H_220 |
||||||
|---|---|---|---|---|---|---|---|---|
| Acc | MCC | Sn | Sp | Acc | MCC | Sn | Sp | |
| BE | 60.10 | 0.20 | 58.79 | 61.41 | 66.50 | 0.33 | 64.00 | 69.00 |
| Mismatch (3) | 60.81 | 0.22 | 57.37 | 64.24 | 59.50 | 0.19 | 58.00 | 61.00 |
| EIIP | 57.37 | 0.15 | 54.55 | 60.20 | 58.00 | 0.16 | 56.00 | 60.00 |
| PseEIIP | 58.99 | 0.18 | 54.75 | 63.23 | 58.00 | 0.16 | 55.00 | 61.00 |
| BE | 60.10 | 0.20 | 58.79 | 61.41 | 66.50 | 0.33 | 64.00 | 69.00 |
| BPB+Kmer(3)+EIIPa | 63.33 | 0.27 | 62.63 | 64.04 | 75.00 | 0.51 | 81.00 | 69.00 |
| BPB+Kmer(3)+PseEIIP | 63.13 | 0.26 | 61.01 | 65.25 | 70.50 | 0.43 | 82.00 | 59.00 |
| BPB+Kmer(3)+BE | 60.91 | 0.22 | 58.99 | 62.83 | 68.00 | 0.36 | 69.00 | 67.00 |
| BPB+Kmer(3)+mismatch(3) | 61.11 | 0.22 | 56.77 | 65.45 | 60.20 | 0.20 | 61.00 | 59.41 |
| BPB+Kmer(3)+EIIP+mismatch(3) | 61.21 | 0.23 | 56.97 | 65.45 | 60.20 | 0.20 | 61.00 | 59.41 |
All values in this row indicate performance with maximum accuracy.
Conclusions
Ψ identification plays an important role in academic research and drug development. In this study, we first extracted the RNA features using the BPB method59,67, 68, 69 for Ψ site prediction, which gives the RNA sequence information from both positive and negative training samples. The evaluated accuracies using the SVM method are 3.55% and 5.09% lower than the iPseU-CUU35 for the H_990 and S_628 datasets. Almost the same results and 5.0% improvement were obtained for M_994, H_200, and S_200, respectively. Then, we combined three kinds of features—Kmer, PC-PseDNC-General, and NCP+ND, where the useful features were further selected by the MRMD method.62 The final accuracies of the combined features using the SVM classifier were achieved at 70.54% and 72.45% for S_628 and M_994, respectively. The predicted Acc of independent S_200 was also improved from 69.0% (BPB features alone) to 77.0% (combined features).
It can be concluded that there are some improvements for the S. cerevisiae and M. musculus using the combined features by the SVM classifier. However, including the six existing predictors, the general accuracies are still 60%–70%, which needs to be further improved for biologist usage. It is clearly known that many kinds of feature extraction methods have been applied to encode RNA sequences to identify Ψ modification, including BPB, Kmer, PC-PseDNC-General, NCP, ND, Mismatch, subsequence, ENAC, EIIP, PseEIIP, BE, DAC, DCC, and DACC in this paper, as well as PSNP, PSDP, and so forth. In addition, many machine-learning-based computational methods70, 71, 72 for the identification of many types of RNA methylations have shown excellent performance (with an Acc of approximately 90%), including m6A, m5C, m1A, and so forth. Thus, the researchers need to reconsider whether there is any sequence feature in the RNA Ψ modification prediction problem. There may be other methods to identify Ψ modification sites that have better performance.
Materials and Methods
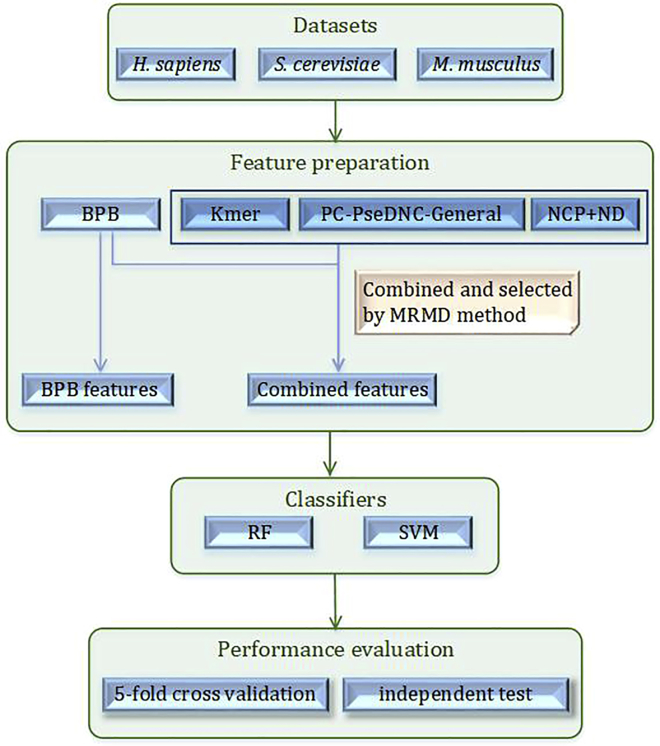
In this study, we use the datasets built by Chen et al.33 from RMBase,73 including training datasets H_990, S_628, and M_990, and independent testing datasets H_200 and S_200 for H. sapiens, S. cerevisiae, and M. musculus, respectively. Here, the BPB features alone as well as the combination of three other kinds of features (Kmer, PC-PseDNC-General, and NCP+ND) using the MRMD method are prepared. Then, two classifiers, RF and SVM, are used separately for model construction. The schematic flowchart of this work is shown in Figure 1.
Figure 1.
Flowchart of Constructed Predictors for Ψ Identification Using the BPB Features
Feature Extraction Methods
BPB
BPB is an effective feature extraction approach that has been successfully applied in bioinformatics with good performance.59,67, 68, 69,74, 75, 76, 77 It can obtain comprehensive sequence information from not only positive but also negative RNA samples. Considering the RNA sequence , the associated BPB feature vector is written as
| (Equation 1) |
where and represent the corresponding nucleotide frequency at each position in positive and negative datasets, respectively. Thus, the BPB features for model training can well reflect the positive and negative position-specific information.
Kmer
Kmer is a common method used to give RNA sequence information, where the feature vector is obtained from the frequencies of k-neighboring nucleotides.60,66 The Kmer features are available at the powerful web server Pse-in-one (http://bioinformatics.hitsz.edu.cn/Pse-in-One/RNA/Kmer/).
PC-PseDNC-General
Similarly, the PC-PseDNC-General features63,64,78 can also be obtained at Pse-in-one (http://bioinformatics.hitsz.edu.cn/Pse-in-One/RNA/PC-PseDNC-General/), where 22 alternative physicochemical properties are provided to generate the pseudo-dinucleotide composition. The corresponding RNA features can be written as:
| (Equation 2) |
with
| (Equation 3) |
Here, indicates the normalized occurrence frequency of the 16 dinucleotides; is the weight factor; and is the j-tier correlation factor demonstrating the sequence-order correlations between all of the most contiguous dinucleotides along a given RNA sequence, where parameter λ gives the highest counted rank (or tier). It can be further expressed as:
| (Equation 4) |
where C(Di, Di+j) is called the correlation function formulated as
| (Equation 5) |
Here, u indicates the number of physicochemical properties investigated and and are the associated values of the gth property for the dinucleotides at position and at , respectively.
NCP+ND
In the iRNA-PseU method, the feature vectors are obtained by incorporating three NCPs (ring structure, hydrogen bond, and functional group) and accumulated occurrence frequency.33 The related chemical properties are described as follows: A and G purines with two rings encoded as 1; C and U pyrimidines with one ring, as 0; the strong hydrogen bonds formed between C and G, as 1 when constructing secondary structures; the weak hydrogen bonds between A and U, as 0; the amino groups A and C, as 1; and the keto groups G and U, as 0. Then, the four nucleotides A, C, G, and U can be encoded as (1, 1, 1), (0, 0, 1), (1, 0, 0), and (0, 1, 0), respectively. In addition, the nucleotide density di is defined as
| (Equation 6) |
where |Ni| is the length of the ith prefix string . Finally, the RNA sequence can be simply represented by a 4l-dimensional vector according to the formulation of PseKNC.
Classifiers and Cross-Validation
RF
RF is a widely used algorithm in prediction problems that effectively combines ensemble tree-structured classifiers.79, 80, 81, 82, 83, 84, 85, 86, 87 It is usually applied to research with a very large number of feature vectors. This classifier consists of hundreds of decision trees, and the final prediction is obtained by major votes. In this article, we used the RF method implemented on the Weka data mining suite with the default parameters for analysis.88
SVM
SVM is a successful machine learning algorithm based on statistical learning theory,89, 90, 91, 92, 93, 94, 95, 96 which has been widely applied in bioinformatics and computational biology.90,97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107, 108 In this method, the original input data are transformed into a higher dimensional feature space (Hilbert space), and then the optimal separating hyperplanes are determined. Here, the LIBSVM package v.3.21109 was used to implement the SVM, where the radial basis kernel function (RBF) was chosen to obtain the best classification hyperplane. The related regularization parameter C and kernel width γ were determined through the optimization procedure, using the default grid search approach written as:
| (Equation 7) |
5-Fold Cross-Validation
Although the jackknife test is effective and stable and has been applied in the iRNA-PseU33 and PseUI,34 it is a very time-consuming process. On the other hand, the predictor iPseU-CUU35 uses 5-fold cross-validation to evaluate performance. Therefore, we chose 5-fold cross-validation on the benchmark datasets for a convenient comparison. Specifically, the benchmark datasets are equally divided into five subsets separately. Then, the four subsets are used to train the model and the remaining one to test. This process is repeated five times when all subsets are applied once for testing. The final performances are an average value of all five testing experiments.110
MRMD
Feature selection aims to select a subset of features by removing redundancy and keeping the most discriminative features.111, 112, 113, 114 MRMD62 is an effective feature selection method to reduce dimensionalities of feature vectors, where the Acc and stability of feature ranking and prediction tasks are both considered. As Xu et al.’s115 related work shows, the performances are improved based on the selected features using the MRMD method. In this method, the features with the maximum relevance and distance are selected as the ultimate sub-feature set for experiments.
Evaluation Parameters
The performance of the constructed models is frequently evaluated using Sn, Sp, Acc, and Matthews correlation coefficient (MCC), which are expressed as:116, 117, 118, 119, 120, 121
| (Equation 8) |
where N+ and N− represent the total number of positive and negative RNA samples considered, in which the incorrectly predicted samples are indicated by and , respectively.
Author Contributions
L.X. and H.X. conceived the idea and designed the overall research. L.D. constructed the predictors, evaluated the performance, and drafted the manuscript. X.L. and H.D. helped to revise the paper; Both authors read, critically revised and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the Natural Science Foundation of China (no. 61902259), the Natural Science Foundation of Guangdong Province (grant no. 2018A0303130084), and the Scientific Research Foundation in Shenzhen (JCYJ20170818100431895, JCYJ20180305163701198, and JCYJ20180306172207178).
Contributor Information
Lei Xu, Email: csleixu@szpt.edu.cn.
Huaikun Xiang, Email: xianghuaikun@szpt.edu.cn.
References
- 1.Hudson G.A., Bloomingdale R.J., Znosko B.M. Thermodynamic contribution and nearest-neighbor parameters of pseudouridine-adenosine base pairs in oligoribonucleotides. RNA. 2013;19:1474–1482. doi: 10.1261/rna.039610.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sloan K.E., Warda A.S., Sharma S., Entian K.D., Lafontaine D.L.J., Bohnsack M.T. Tuning the ribosome: The influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 2017;14:1138–1152. doi: 10.1080/15476286.2016.1259781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ge J., Yu Y.T. RNA pseudouridylation: new insights into an old modification. Trends Biochem. Sci. 2013;38:210–218. doi: 10.1016/j.tibs.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han S., Liang Y., Ma Q., Xu Y., Zhang Y., Du W., Wang C., Li Y. LncFinder: an integrated platform for long non-coding RNA identification utilizing sequence intrinsic composition, structural information and physicochemical property. Brief. Bioinform. 2018 doi: 10.1093/bib/bby065. Published online July 31, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu S.J., Xie J., Li Y., Yu B., Ma Q., Liu B.Q. Identification of lncRNAs-gene interactions in transcription regulation based on co-expression analysis of RNA-seq data. Math. Biosci. Eng. 2019;16:7112–7125. doi: 10.3934/mbe.2019357. [DOI] [PubMed] [Google Scholar]
- 6.Cantara W.A., Crain P.F., Rozenski J., McCloskey J.A., Harris K.A., Zhang X., Vendeix F.A., Fabris D., Agris P.F. The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res. 2011;39:D195–D201. doi: 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boccaletto P., Machnicka M.A., Purta E., Piatkowski P., Baginski B., Wirecki T.K., de Crécy-Lagard V., Ross R., Limbach P.A., Kotter A. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46(D1):D303–D307. doi: 10.1093/nar/gkx1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang J., Fu J., Wang Y., Luo Y., Yang Q., Li B., Tu G., Hong J., Cui X., Chen Y. Simultaneous improvement in the precision, accuracy, and robustness of label-free proteome quantification by optimizing data manipulation chains. Mol. Cell. Proteomics. 2019;18:1683–1699. doi: 10.1074/mcp.RA118.001169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng L., Wang P., Tian R., Wang S., Guo Q., Luo M., Zhou W., Liu G., Jiang H., Jiang Q. LncRNA2Target v2.0: a comprehensive database for target genes of lncRNAs in human and mouse. Nucleic Acids Res. 2019;47(D1):D140–D144. doi: 10.1093/nar/gky1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng L., Sun J., Xu W., Dong L., Hu Y., Zhou M. OAHG: an integrated resource for annotating human genes with multi-level ontologies. Sci. Rep. 2016;6:34820. doi: 10.1038/srep34820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J., Peng H., Han G., Cai H., Cai J. HOGMMNC: a higher order graph matching with multiple network constraints model for gene-drug regulatory modules identification. Bioinformatics. 2019;35:602–610. doi: 10.1093/bioinformatics/bty662. [DOI] [PubMed] [Google Scholar]
- 12.Charette M., Gray M.W. Pseudouridine in RNA: what, where, how, and why. IUBMB Life. 2000;49:341–351. doi: 10.1080/152165400410182. [DOI] [PubMed] [Google Scholar]
- 13.Carlile T.M., Rojas-Duran M.F., Zinshteyn B., Shin H., Bartoli K.M., Gilbert W.V. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature. 2014;515:143–146. doi: 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lovejoy A.F., Riordan D.P., Brown P.O. Transcriptome-wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS ONE. 2014;9:e110799. doi: 10.1371/journal.pone.0110799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwartz S., Bernstein D.A., Mumbach M.R., Jovanovic M., Herbst R.H., León-Ricardo B.X., Engreitz J.M., Guttman M., Satija R., Lander E.S. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014;159:148–162. doi: 10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X., Zhu P., Ma S., Song J., Bai J., Sun F., Yi C. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat. Chem. Biol. 2015;11:592–597. doi: 10.1038/nchembio.1836. [DOI] [PubMed] [Google Scholar]
- 17.Tang J., Fu J., Wang Y., Li B., Li Y., Yang Q., Cui X., Hong J., Li X., Chen Y. ANPELA: analysis and performance assessment of the label-free quantification workflow for metaproteomic studies. Brief. Bioinform. 2019 doi: 10.1093/bib/bby127. Published online January 15, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou M., Zhao H., Wang X., Sun J., Su J. Analysis of long noncoding RNAs highlights region-specific altered expression patterns and diagnostic roles in Alzheimer’s disease. Brief. Bioinform. 2019;20:598–608. doi: 10.1093/bib/bby021. [DOI] [PubMed] [Google Scholar]
- 19.Zhou M., Zhang Z., Zhao H., Bao S., Cheng L., Sun J. An immune-related six-lncRNA signature to improve prognosis prediction of glioblastoma multiforme. Mol. Neurobiol. 2018;55:3684–3697. doi: 10.1007/s12035-017-0572-9. [DOI] [PubMed] [Google Scholar]
- 20.Zhou M., Hu L., Zhang Z., Wu N., Sun J., Su J. Recurrence-associated long non-coding RNA signature for determining the risk of recurrence in patients with colon cancer. Mol. Ther. Nucleic Acids. 2018;12:518–529. doi: 10.1016/j.omtn.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou M., Zhao H., Xu W., Bao S., Cheng L., Sun J. Discovery and validation of immune-associated long non-coding RNA biomarkers associated with clinically molecular subtype and prognosis in diffuse large B cell lymphoma. Mol. Cancer. 2017;16 doi: 10.1186/s12943-017-0580-4. Article 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou M., Zhao H., Wang Z., Cheng L., Yang L., Shi H., Yang H., Sun J. Identification and validation of potential prognostic lncRNA biomarkers for predicting survival in patients with multiple myeloma. J. Exp. Clin. Cancer Res. 2015;34:102. doi: 10.1186/s13046-015-0219-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu L., Zhao J., Gao L. Predicting potential drugs for breast cancer based on miRNA and tissue specificity. Int. J. Biol. Sci. 2018;14:971–982. doi: 10.7150/ijbs.23350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang G., Shi J., Wu W., Yue X., Zhang W. Sequence-based bacterial small RNAs prediction using ensemble learning strategies. BMC Bioinformatics. 2018;19(Suppl. 20):503. doi: 10.1186/s12859-018-2535-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang W., Qu Q., Zhang Y., Wang W. The linear neighborhood propagation method for predicting long non-coding RNA–protein interactions. Neurocomputing. 2018;273:526–534. [Google Scholar]
- 26.Zhang W., Yue X., Tang G., Wu W., Huang F., Zhang X. SFPEL-LPI: sequence-based feature projection ensemble learning for predicting lncRNA-protein interactions. PLoS Comput. Biol. 2018;14:e1006616. doi: 10.1371/journal.pcbi.1006616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang W., Li Z., Guo W., Yang W., Huang F. A fast linear neighborhood similarity-based network link inference method to predict microRNA-disease associations. IEEE/ACM Trans. Comput. Biol. Bioinform. 2019 doi: 10.1109/TCBB.2019.2931546. Published online July 29, 2019. [DOI] [PubMed] [Google Scholar]
- 28.Li D., Luo L., Zhang W., Liu F., Luo F. A genetic algorithm-based weighted ensemble method for predicting transposon-derived piRNAs. BMC Bioinformatics. 2016;17:329. doi: 10.1186/s12859-016-1206-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao Z., Li D., Wang X., Li L., Zou Q. Cancer diagnosis from isomiR expression with machine learning method. Curr. Bioinform. 2018;13:57–63. [Google Scholar]
- 30.Xu A., Chen J., Peng H., Han G., Cai H. Simultaneous interrogation of cancer omics to identify subtypes with significant clinical differences. Front. Genet. 2019;10:236. doi: 10.3389/fgene.2019.00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Panwar B., Raghava G.P.S. Prediction of uridine modifications in tRNA sequences. BMC Bioinformatics. 2014;15:326. doi: 10.1186/1471-2105-15-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y.H., Zhang G., Cui Q. PPUS: a web server to predict PUS-specific pseudouridine sites. Bioinformatics. 2015;31:3362–3364. doi: 10.1093/bioinformatics/btv366. [DOI] [PubMed] [Google Scholar]
- 33.Chen W., Tang H., Ye J., Lin H., Chou K.C. iRNA-PseU: identifying RNA pseudouridine sites. Mol. Ther. Nucleic Acids. 2016;5:e332. doi: 10.1038/mtna.2016.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He J., Fang T., Zhang Z., Huang B., Zhu X., Xiong Y. PseUI: pseudouridine sites identification based on RNA sequence information. BMC Bioinformatics. 2018;19:306. doi: 10.1186/s12859-018-2321-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tahir M., Tayara H., Chong K.T. iPseU-CNN: identifying RNA pseudouridine sites using convolutional neural networks. Mol. Ther. Nucleic Acids. 2019;16:463–470. doi: 10.1016/j.omtn.2019.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu K., Chen W., Lin H. XG-PseU: an eXtreme Gradient Boosting based method for identifying pseudouridine sites. Mol. Genet. Genomics. 2019 doi: 10.1007/s00438-019-01600-9. Published online August 7, 2019. [DOI] [PubMed] [Google Scholar]
- 37.Chen C.Y., Chuang T.J. Comment on “A comprehensive overview and evaluation of circular RNA detection tools”. PLoS Comput. Biol. 2019;15:e1006158. doi: 10.1371/journal.pcbi.1006158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xin Z., Ma Q., Ren S., Wang G., Li F. The understanding of circular RNAs as special triggers in carcinogenesis. Brief. Funct. Genomics. 2017;16:80–86. doi: 10.1093/bfgp/elw001. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Z., Zhao Y., Liao X., Shi W., Li K., Zou Q., Peng S. Deep learning in omics: a survey and guideline. Brief. Funct. Genomics. 2019;18:41–57. doi: 10.1093/bfgp/ely030. [DOI] [PubMed] [Google Scholar]
- 40.Wei L., Su R., Wang B., Li X., Zou Q., Gao X. Integration of deep feature representations and handcrafted features to improve the prediction of N 6-methyladenosine sites. Neurocomputing. 2019;324:3–9. [Google Scholar]
- 41.Lv Z., Ao C., Zou Q. Protein function prediction: from traditional classifier to deep learning. Proteomics. 2019;19:e1900119. doi: 10.1002/pmic.201900119. [DOI] [PubMed] [Google Scholar]
- 42.Wei L., Ding Y., Su R., Tang J., Zou Q. Prediction of human protein subcellular localization using deep learning. J. Parallel Distrib. Comput. 2018;117:212–217. [Google Scholar]
- 43.Chen W., Ding H., Zhou X., Lin H., Chou K.C. iRNA(m6A)-PseDNC: identifying N6-methyladenosine sites using pseudo dinucleotide composition. Anal. Biochem. 2018;561-562:59–65. doi: 10.1016/j.ab.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 44.Chen K., Wei Z., Zhang Q., Wu X., Rong R., Lu Z., Su J., de Magalhães J.P., Rigden D.J., Meng J. WHISTLE: a high-accuracy map of the human N6-methyladenosine (m6A) epitranscriptome predicted using a machine learning approach. Nucleic Acids Res. 2019;47:e41. doi: 10.1093/nar/gkz074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zou Q., Xing P., Wei L., Liu B. Gene2vec: gene subsequence embedding for prediction of mammalian N6-methyladenosine sites from mRNA. RNA. 2019;25:205–218. doi: 10.1261/rna.069112.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feng P., Ding H., Chen W., Lin H. Identifying RNA 5-methylcytosine sites via pseudo nucleotide compositions. Mol. Biosyst. 2016;12:3307–3311. doi: 10.1039/c6mb00471g. [DOI] [PubMed] [Google Scholar]
- 47.Qiu W.R., Jiang S.Y., Xu Z.C., Xiao X., Chou K.C. iRNAm5C-PseDNC: identifying RNA 5-methylcytosine sites by incorporating physical-chemical properties into pseudo dinucleotide composition. Oncotarget. 2017;8:41178–41188. doi: 10.18632/oncotarget.17104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang M., Xu Y., Li L., Liu Z., Yang X., Yu D.J. Accurate RNA 5-methylcytosine site prediction based on heuristic physical-chemical properties reduction and classifier ensemble. Anal. Biochem. 2018;550:41–48. doi: 10.1016/j.ab.2018.03.027. [DOI] [PubMed] [Google Scholar]
- 49.Sabooh M.F., Iqbal N., Khan M., Khan M., Maqbool H.F. Identifying 5-methylcytosine sites in RNA sequence using composite encoding feature into Chou’s PseKNC. J. Theor. Biol. 2018;452:1–9. doi: 10.1016/j.jtbi.2018.04.037. [DOI] [PubMed] [Google Scholar]
- 50.Li J., Huang Y., Yang X., Zhou Y., Zhou Y. RNAm5Cfinder: a web-server for predicting RNA 5-methylcytosine (m5C) sites based on random forest. Sci. Rep. 2018;8:17299. doi: 10.1038/s41598-018-35502-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song J., Zhai J., Bian E., Song Y., Yu J., Ma C. Transcriptome-wide annotation of m5C RNA modifications using machine learning. Front. Plant Sci. 2018;9:519. doi: 10.3389/fpls.2018.00519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lv H., Zhang Z.M., Li S.H., Tan J.X., Chen W., Lin H. Evaluation of different computational methods on 5-methylcytosine sites identification. Brief. Bioinform. 2019:bbz048. doi: 10.1093/bib/bbz048. [DOI] [PubMed] [Google Scholar]
- 53.Xue W., Yang F., Wang P., Zheng G., Chen Y., Yao X., Zhu F. What contributes to serotonin-norepinephrine reuptake inhibitors’ dual-targeting mechanism? The key role of transmembrane domain 6 in human serotonin and norepinephrine transporters revealed by molecular dynamics simulation. ACS Chem. Neurosci. 2018;9:1128–1140. doi: 10.1021/acschemneuro.7b00490. [DOI] [PubMed] [Google Scholar]
- 54.Chen W., Feng P., Tang H., Ding H., Lin H. RAMPred: identifying the N(1)-methyladenosine sites in eukaryotic transcriptomes. Sci. Rep. 2016;6:31080. doi: 10.1038/srep31080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feng P., Ding H., Yang H., Chen W., Lin H., Chou K.-C. iRNA-PseColl: identifying the occurrence sites of different RNA modifications by incorporating collective effects of nucleotides into PseKNC. Mol. Ther. Nucleic Acids. 2017;7:155–163. doi: 10.1016/j.omtn.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen W., Feng P., Yang H., Ding H., Lin H., Chou K.C. iRNA-3typeA: identifying three types of modification at RNA’s adenosine sites. Mol. Ther. Nucleic Acids. 2018;11:468–474. doi: 10.1016/j.omtn.2018.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen X., Sun Y.Z., Liu H., Zhang L., Li J.Q., Meng J. RNA methylation and diseases: experimental results, databases, Web servers and computational models. Brief. Bioinform. 2019;20:896–917. doi: 10.1093/bib/bbx142. [DOI] [PubMed] [Google Scholar]
- 58.Li Y.Z., FY X., FY X. KELMPSP: pseudouridine sites identification based on kernel extreme learning machine. Chin. J. Biochem. Mol. Biol. 2018;34:785–793. [Google Scholar]
- 59.Shao J., Xu D., Tsai S.-N., Wang Y., Ngai S.-M. Computational identification of protein methylation sites through bi-profile Bayes feature extraction. PLoS ONE. 2009;4:e4920. doi: 10.1371/journal.pone.0004920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wei L., Liao M., Gao Y., Ji R., He Z., Zou Q. Improved and promising identification of human microRNAs by incorporating a high-quality negative set. IEEE/ACM Trans. Comput. Biol. Bioinformatics. 2014;11:192–201. doi: 10.1109/TCBB.2013.146. [DOI] [PubMed] [Google Scholar]
- 61.Liu B., Liu F., Wang X., Chen J., Fang L., Chou K.C. Pse-in-One: a web server for generating various modes of pseudo components of DNA, RNA, and protein sequences. Nucleic Acids Res. 2015;43(W1):W65–W71. doi: 10.1093/nar/gkv458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zou Q., Zeng J.C., Cao L.J., Ji R.R. A novel features ranking metric with application to scalable visual and bioinformatics data classification. Neurocomputing. 2016;173:346–354. [Google Scholar]
- 63.Chen W., Zhang X., Brooker J., Lin H., Zhang L., Chou K.C. PseKNC-General: a cross-platform package for generating various modes of pseudo nucleotide compositions. Bioinformatics. 2015;31:119–120. doi: 10.1093/bioinformatics/btu602. [DOI] [PubMed] [Google Scholar]
- 64.Yang H., Lv H., Ding H., Chen W., Lin H. iRNA-2OM: a sequence-based predictor for identifying 2′-O-methylation sites in Homo sapiens. J. Comput. Biol. 2018;25:1266–1277. doi: 10.1089/cmb.2018.0004. [DOI] [PubMed] [Google Scholar]
- 65.Chen Z., Zhao P., Li F., Marquez-Lago T.T., Leier A., Revote J., Zhu Y., Powell D.R., Akutsu T., Webb G.I. iLearn: an integrated platform and meta-learner for feature engineering, machine-learning analysis and modeling of DNA, RNA and protein sequence data. Brief. Bioinform. 2019:bbz041. doi: 10.1093/bib/bbz041. [DOI] [PubMed] [Google Scholar]
- 66.Liu B., Gao X., Zhang H. BioSeq-Analysis2.0: an updated platform for analyzing DNA, RNA and protein sequences at sequence level and residue level based on machine learning approaches. Nucleic Acids Res. 2019;47:e127. doi: 10.1093/nar/gkz740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jia C., Liu T., Chang A.K., Zhai Y. Prediction of mitochondrial proteins of malaria parasite using bi-profile Bayes feature extraction. Biochimie. 2011;93:778–782. doi: 10.1016/j.biochi.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 68.Zhao X., Zhang J., Ning Q., Sun P., Ma Z., Yin M. Identification of protein pupylation sites using bi-profile Bayes feature extraction and ensemble learning. Math. Probl. Eng. 2013;2013:283129. [Google Scholar]
- 69.Jia C.Z., He W.Y., Yao Y.H. OH-PRED: prediction of protein hydroxylation sites by incorporating adapted normal distribution bi-profile Bayes feature extraction and physicochemical properties of amino acids. J. Biomol. Struct. Dyn. 2017;35:829–835. doi: 10.1080/07391102.2016.1163294. [DOI] [PubMed] [Google Scholar]
- 70.Song T., Rodríguez-Patón A., Zheng P., Zeng X. Spiking neural P systems with colored spikes. IEEE Trans. Cogn. Dev. Syst. 2018;10:1106–1115. doi: 10.1109/TNB.2018.2873221. [DOI] [PubMed] [Google Scholar]
- 71.Xu H., Zeng W., Zhang D., Zeng X. MOEA/HD: a multiobjective evolutionary algorithm based on hierarchical decomposition. IEEE Trans. Cybern. 2019;49:517–526. doi: 10.1109/TCYB.2017.2779450. [DOI] [PubMed] [Google Scholar]
- 72.Cabarle F.G.C., Adorna H.N., Jiang M., Zeng X. Spiking neural P systems with scheduled synapses. IEEE Trans. Nanobioscience. 2017;16:792–801. doi: 10.1109/TNB.2017.2762580. [DOI] [PubMed] [Google Scholar]
- 73.Sun W.J., Li J.H., Liu S., Wu J., Zhou H., Qu L.H., Yang J.H. RMBase: a resource for decoding the landscape of RNA modifications from high-throughput sequencing data. Nucleic Acids Res. 2016;44(D1):D259–D265. doi: 10.1093/nar/gkv1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.He W., Jia C., Zou Q. 4mCPred: machine learning methods for DNA N4-methylcytosine sites prediction. Bioinformatics. 2019;35:593–601. doi: 10.1093/bioinformatics/bty668. [DOI] [PubMed] [Google Scholar]
- 75.Li B., Tang J., Yang Q., Li S., Cui X., Li Y., Chen Y., Xue W., Li X., Zhu F. NOREVA: normalization and evaluation of MS-based metabolomics data. Nucleic Acids Res. 2017;45(W1):W162–W170. doi: 10.1093/nar/gkx449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cheng L., Hu Y., Sun J., Zhou M., Jiang Q. DincRNA: a comprehensive web-based bioinformatics toolkit for exploring disease associations and ncRNA function. Bioinformatics. 2018;34:1953–1956. doi: 10.1093/bioinformatics/bty002. [DOI] [PubMed] [Google Scholar]
- 77.Cheng L., Jiang Y., Ju H., Sun J., Peng J., Zhou M., Hu Y. InfAcrOnt: calculating cross-ontology term similarities using information flow by a random walk. BMC Genomics. 2018;19(Suppl. 1):919. doi: 10.1186/s12864-017-4338-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu B. BioSeq-Analysis: a platform for DNA, RNA and protein sequence analysis based on machine learning approaches. Brief. Bioinform. 2017;20:1280–1294. doi: 10.1093/bib/bbx165. [DOI] [PubMed] [Google Scholar]
- 79.Breiman L. Random forests. Mach. Learn. 2001;45:5–32. [Google Scholar]
- 80.Li Y., Shi X., Liang Y., Xie J., Zhang Y., Ma Q. RNA-TVcurve: a web server for RNA secondary structure comparison based on a multi-scale similarity of its triple vector curve representation. BMC Bioinformatics. 2017;18:51. doi: 10.1186/s12859-017-1481-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu B., Yang F., Huang D.S., Chou K.C. iPromoter-2L: a two-layer predictor for identifying promoters and their types by multi-window-based PseKNC. Bioinformatics. 2018;34:33–40. doi: 10.1093/bioinformatics/btx579. [DOI] [PubMed] [Google Scholar]
- 82.Ding Y., Tang J., Guo F. Identification of protein-protein interactions via a novel matrix-based sequence representation model with amino acid contact information. Int. J. Mol. Sci. 2016;17:E1623. doi: 10.3390/ijms17101623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ding Y., Tang J., Guo F. Predicting protein-protein interactions via multivariate mutual information of protein sequences. BMC Bioinformatics. 2016;17:398. doi: 10.1186/s12859-016-1253-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yu L., Su R., Wang B., Zhang L., Zou Y., Zhang J., Gao L. Prediction of novel drugs for hepatocellular carcinoma based on multi-source random walk. IEEE/ACM Trans. Comput. Biol. Bioinformatics. 2017;14:966–977. doi: 10.1109/TCBB.2016.2550453. [DOI] [PubMed] [Google Scholar]
- 85.Ru X., Li L., Zou Q. Incorporating distance-based Top-n-gram and random forest to identify electron transport proteins. J. Proteome Res. 2019;18:2931–2939. doi: 10.1021/acs.jproteome.9b00250. [DOI] [PubMed] [Google Scholar]
- 86.Su R., Liu X., Wei L., Zou Q. Deep-Resp-Forest: a deep forest model to predict anti-cancer drug response. Methods. 2019;166:91–102. doi: 10.1016/j.ymeth.2019.02.009. [DOI] [PubMed] [Google Scholar]
- 87.Xu L., Liang G., Liao C., Chen G.D., Chang C.C. k-skip-n-gram-RF: a random forest based method for Alzheimer’s Disease protein identification. Front. Genet. 2019;10:33. doi: 10.3389/fgene.2019.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Frank E., Hall M., Trigg L., Holmes G., Witten I.H. Data mining in bioinformatics using Weka. Bioinformatics. 2004;20:2479–2481. doi: 10.1093/bioinformatics/bth261. [DOI] [PubMed] [Google Scholar]
- 89.Cortes C., Vapnik V. Support-vector networks. Mach. Learn. 1995;20:273–297. [Google Scholar]
- 90.Nello Cristianini J.S.-T. Cambridge University Press; 2000. An Introduction of Support Vector Machines and Other Kernel-Based Learning Methods. [Google Scholar]
- 91.Zhang X., Zou Q., Rodriguez-Paton A., Zeng X. Meta-path methods for prioritizing candidate disease miRNAs. IEEE/ACM Trans. Comput. Biol. Bioinformatics. 2019;16:283–291. doi: 10.1109/TCBB.2017.2776280. [DOI] [PubMed] [Google Scholar]
- 92.Zou Q., Li J., Song L., Zeng X., Wang G. Similarity computation strategies in the microRNA-disease network: a survey. Brief. Funct. Genomics. 2016;15:55–64. doi: 10.1093/bfgp/elv024. [DOI] [PubMed] [Google Scholar]
- 93.Zeng X., Liao Y., Liu Y., Zou Q. Prediction and validation of disease genes using HeteSim scores. IEEE/ACM Trans. Comput. Biol. Bioinformatics. 2017;14:687–695. doi: 10.1109/TCBB.2016.2520947. [DOI] [PubMed] [Google Scholar]
- 94.Sun Y., Xiong Y., Xu Q., Wei D. A hadoop-based method to predict potential effective drug combination. BioMed Res. Int. 2014;2014:196858. doi: 10.1155/2014/196858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xu L., Liang G., Liao C., Chen G.-D., Chang C.-C. An efficient classifier for Alzheimer’s Disease genes identification. Molecules. 2018;23:3140. doi: 10.3390/molecules23123140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xu L., Liang G., Shi S., Liao C. SeqSVM: a sequence-based support vector machine method for identifying antioxidant proteins. Int. J. Mol. Sci. 2018;19:E17773. doi: 10.3390/ijms19061773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chou K.C., Cai Y.D. Using functional domain composition and support vector machines for prediction of protein subcellular location. J. Biol. Chem. 2002;277:45765–45769. doi: 10.1074/jbc.M204161200. [DOI] [PubMed] [Google Scholar]
- 98.Cai Y.D., Zhou G.P., Chou K.C. Support vector machines for predicting membrane protein types by using functional domain composition. Biophys. J. 2003;84:3257–3263. doi: 10.1016/S0006-3495(03)70050-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhu X.J., Feng C.Q., Lai H.Y., Chen W., Lin H. Predicting protein structural classes for low-similarity sequences by evaluating different features. Knowl. Based Syst. 2019;163:787–793. [Google Scholar]
- 100.Chen W., Feng P., Liu T., Jin D. Recent advances in machine learning methods for predicting heat shock proteins. Curr. Drug Metab. 2019;20:224–228. doi: 10.2174/1389200219666181031105916. [DOI] [PubMed] [Google Scholar]
- 101.Li Y.H., Li X.X., Hong J.J., Wang Y.X., Fu J.B., Yang H., Yu C.Y., Li F.C., Hu J., Xue W.W. Clinical trials, progression-speed differentiating features and swiftness rule of the innovative targets of first-in-class drugs. Brief. Bioinform. 2019 doi: 10.1093/bib/bby130. Published January 23, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu B., Li K. iPromoter-2L2.0: identifying promoters and their types by combining smoothing cutting window algorithm and sequence-based features. Mol. Ther. Nucleic Acids. 2019;18:80–87. doi: 10.1016/j.omtn.2019.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu B., Li C.C., Yan K. DeepSVM-fold: protein fold recognition by combining support vector machines and pairwise sequence similarity scores generated by deep learning networks. Brief. Bioinform. 2019 doi: 10.1093/bib/bbz098. Published online October 28, 2019. [DOI] [PubMed] [Google Scholar]
- 104.Ding Y., Tang J., Guo F. Identification of drug-target interactions via multiple information integration. Inf. Sci. 2017;418-419:546–560. [Google Scholar]
- 105.Xiong Y., Qiao Y., Kihara D., Zhang H.Y., Zhu X., Wei D.Q. Survey of machine learning techniques for prediction of the isoform specificity of cytochrome P450 substrates. Curr. Drug Metab. 2019;20:229–235. doi: 10.2174/1389200219666181019094526. [DOI] [PubMed] [Google Scholar]
- 106.Xiong Y., Liu J., Wei D.Q. An accurate feature-based method for identifying DNA-binding residues on protein surfaces. Proteins. 2011;79:509–517. doi: 10.1002/prot.22898. [DOI] [PubMed] [Google Scholar]
- 107.Wei L., Xing P., Zeng J., Chen J., Su R., Guo F. Improved prediction of protein-protein interactions using novel negative samples, features, and an ensemble classifier. Artif. Intell. Med. 2017;83:67–74. doi: 10.1016/j.artmed.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 108.Wei L., Wan S., Guo J., Wong K.K. A novel hierarchical selective ensemble classifier with bioinformatics application. Artif. Intell. Med. 2017;83:82–90. doi: 10.1016/j.artmed.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 109.Chang C.C., Lin C.J. LIBSVM: a library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011;2 Article 27. [Google Scholar]
- 110.Chou K.C., Zhang C.T. Prediction of protein structural classes. Crit. Rev. Biochem. Mol. Biol. 1995;30:275–349. doi: 10.3109/10409239509083488. [DOI] [PubMed] [Google Scholar]
- 111.Zhu P.F., Xu Q., Hu Q.H., Zhang C.Q. Co-regularized unsupervised feature selection. Neurocomputing. 2018;275:2855–2863. [Google Scholar]
- 112.Zhu P.F., Xu Q., Hu Q.H., Zhang C.Q., Zhao H. Multi-label feature selection with missing labels. Pattern Recognit. 2018;74:488–502. [Google Scholar]
- 113.Zhu P.F., Zhu W.C., Hu Q.H., Zhang C.Q., Zuo W.M. Subspace clustering guided unsupervised feature selection. Pattern Recognit. 2017;66:364–374. [Google Scholar]
- 114.Yu L., Yao S., Gao L., Zha Y. Conserved disease modules extracted from multilayer heterogeneous disease and gene networks for understanding disease mechanisms and predicting disease treatments. Front. Genet. 2019;9:745. doi: 10.3389/fgene.2018.00745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Xu L., Liang G., Wang L., Liao C. A novel hybrid sequence-based model for identifying anticancer peptides. Genes (Basel) 2018;9:158. doi: 10.3390/genes9030158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chen W., Feng P.M., Lin H., Chou K.C. iRSpot-PseDNC: identify recombination spots with pseudo dinucleotide composition. Nucleic Acids Res. 2013;41:e68. doi: 10.1093/nar/gks1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Xu Y., Ding J., Wu L.-Y., Chou K.-C. iSNO-PseAAC: predict cysteine S-nitrosylation sites in proteins by incorporating position specific amino acid propensity into pseudo amino acid composition. PLoS ONE. 2013;8:e55844. doi: 10.1371/journal.pone.0055844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chen W., Lv H., Nie F., Lin H. i6mA-Pred: identifying DNA N6-methyladenine sites in the rice genome. Bioinformatics. 2019;35:2796–2800. doi: 10.1093/bioinformatics/btz015. [DOI] [PubMed] [Google Scholar]
- 119.Ding Y., Tang J., Guo F. Identification of drug-side effect association via semi-supervised model and multiple kernel learning. IEEE J. Biomed. Health Inform. 2019;23:2619–2632. doi: 10.1109/JBHI.2018.2883834. [DOI] [PubMed] [Google Scholar]
- 120.Ding Y., Tang J., Guo F. Identification of drug-side effect association via multiple information integration with centered kernel alignment. Neurocomputing. 2019;325:211–224. [Google Scholar]
- 121.Shen Y., Tang J., Guo F. Identification of protein subcellular localization via integrating evolutionary and physicochemical information into Chou’s general PseAAC. J. Theor. Biol. 2019;462:230–239. doi: 10.1016/j.jtbi.2018.11.012. [DOI] [PubMed] [Google Scholar]