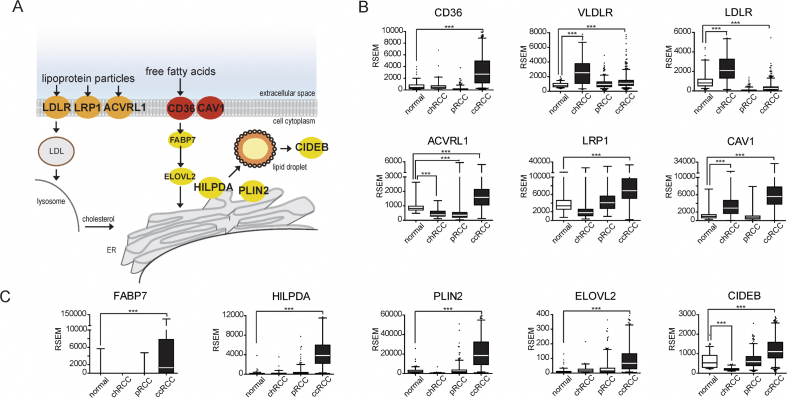
Figure 4.
mRNA levels of genes involved in lipid uptake and storage in human kidney cancer. Transcriptomics data obtained by the TCGA from normal human kidney tissue (n = 129), chromophobe (chRCC, n = 66), papillary (pRCC, n = 290), and clear cell renal cell carcinoma (ccRCC, n = 533) specimens were analyzed for lipid uptake and storage genes. A illustrates some of the lipid uptake (orange/red) and storage (yellow) pathways. B illustrates the quantitative mRNA levels (RNA-Seq by Expectation Maximization [RSEM]) of lipid uptake genes in normal tissue and different subtypes of kidney cancer, respectively. Note statistically significant increases in transcripts of lipid uptake genes CD36, VLDLR, ACVRL1, LRP1, and CAV1 in human ccRCC compared with the normal kidney. As shown in C, mRNA levels of lipid storage genes FABP7, HILPDA, PLIN2, ELOVL2, and CIDEB are also increased in ccRCC. Together, these data indicate that increased transcripts of genes involved in both dietary lipid uptake and lipid storage are common in ccRCC compared with both normal kidney and with other histologic subtypes of RCC. Statistical significance of differences in cancer specimens as compared with the normal kidney tissue was assessed using ANOVA (p < 0.05).