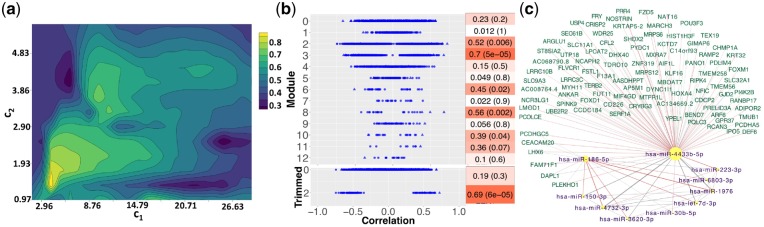
Fig. 2.
FEV1pp results. (a) Total prediction error contour. The x- and y-axes indicate LASSO penalties considered for mRNA and miRNA, respectively. Blue to yellow scale indicates increasing of total prediction error. (b) Correlations with FEV1pp for full (top) and trimmed (bottom) modules. The mRNAs are circles and the miRNAs are triangles. All features excluded in identified modules are grouped as Module 0. Heatmap indicates module–trait correlations (with P-values in parentheses). (c) Trimmed module network. Negative edge is in red; positive edges are in gray. Width of the network edges indicates the connection strength. Yellow nodes with purple labels are miRNAs; magenta nodes with green labels are mRNAs. Node size corresponds to the number of edges connections