Fig. 1.
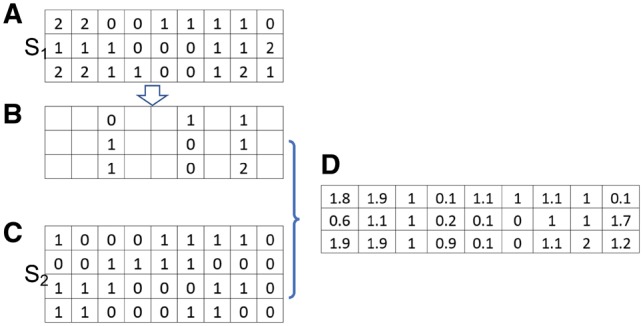
Estimate correlation of imputed SNPs in the validation GWAS dataset. The subjects in The 1000 Genome Project with relevant ancestry are divided as two sets S1 and S2. (A) The genotype of the subjects in S1. (B) Only SNPs that are genotyped in the validation GWAS dataset are kept. (C) The haplotypes in S2 are used as reference panel for imputation. (D) Imputation is performed to derive the genotypic dosage for SNPs that are not genotyped in the validation GWAS dataset. The correlation between two SNPs is calculated based on the imputed genotypic dosages
