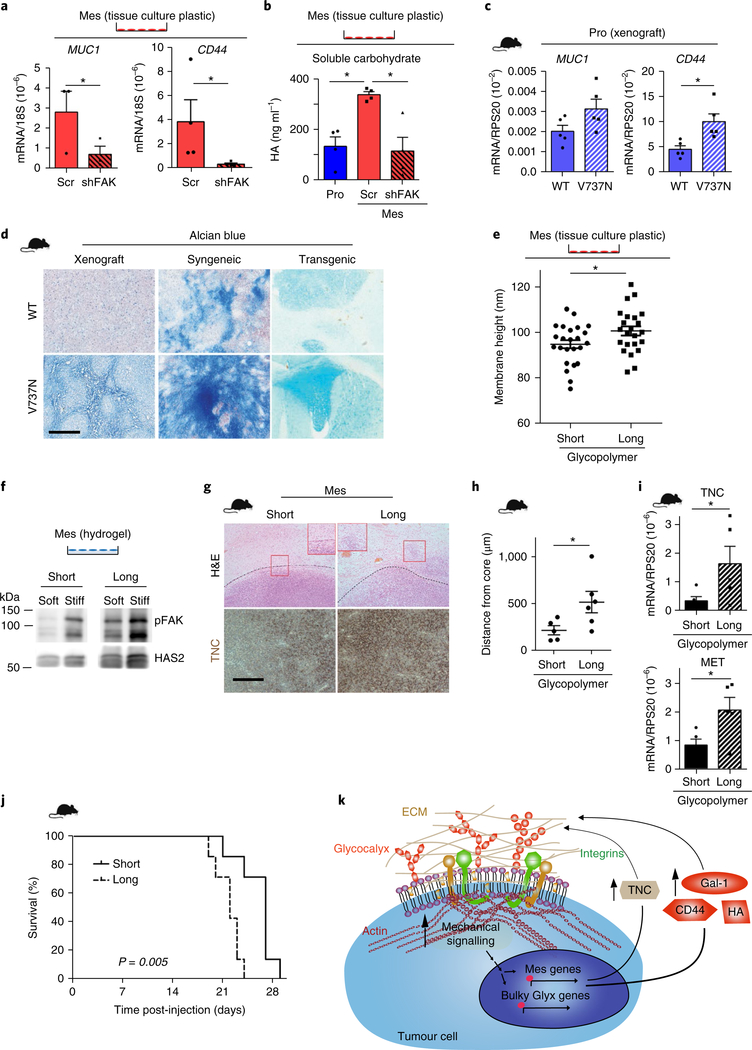
Fig. 5 |. Glycocalyx bulkiness is fostered by enhanced mechanics and is associated with GBM aggression.
a, RNA from control (Scr) and mesenchymal shFAK cells was analysed by qPCR. Mean ± s.e.m. (n = 3 and 4 independent experiments for MUC1 and CD44, respectively). *P = 0.03 and 0.027 for MUC1 and CD44, respectively, by one-sided paired t-test. b, Hyaluronan (HA) was measured from human proneural (Pro), Scr and shFAK cells. Mean ± s.e.m. (n = 4 independent experiments). *P = 0.01 and 0.021 for proneural verus Scr and Scr versus shFAK, respectively, by two-sided paired t-test. c, qPCR of V737N and WT proneural xenografts. Mean ± s.e.m. (n = 5 mice per group). *P = 0.08 and 0.012 for MUC1 and CD44, respectively, by two-sided unpaired t-test. d, Alcian blue staining of WT and V737N xenograft, syngeneic and transgenic proneural tumour models. Scale bar, 250 μm (n = 3 mice per group). e, Mesenchymal cells were loaded with short (3 nm) or long (90 nm) glycopolymers, and glycocalyx height was measured by SAIM. Mean ± s.e.m.; each point represents measurement of glycocalyx height per frame (n = 24 and 23 measurements for short and long glycopolymers, respectively, pooled from two independent experiments). *P = 0.035 by two-sided paired t-test. f, Immunoblot assays were performed for pY397-FAK and HAS2 on soft and stiff hydrogels for long versus short glycopolymer decorated mesenchymal cells (n = 3 independent experiments). g, Cells loaded with long or short glycopolymers were injected into nude mice, and xenografts stained for TNC and by H&E. Insets show differential invasion and black broken lines indicate tumour boundary (n = 7 mice per group). Scale bar, 250 μm. h, Quantification of invasive edge was performed as for Fig. 3j. Mean ± s.e.m. (n = 5 and 6 distance measurements for WT and V737N tumours, respectively, for 4 tissue samples per group). *P = 0.047 by two-sided unpaired t-test. i, qPCR was performed for these tumours. Mean ± s.e.m. (n = 5 tissues per group). *P = 0.048 and 0.028 for TNC and MET, respectively, by one-sided unpaired t-test. j, Survival analysed by Kaplan–Meier test (n = 7 mice per group). P value calculated using two-sided logrank test. k, Schematic showing that enhanced mechanical signalling in tumour cells promotes the upregulation of mesenchymal and bulky glycocalyx (Glyx) genes, which results in the production of ECM proteins such as TNC and bulky glycocalyx components and modulators (CD44, hyaluronan (HA) and Gal-1. Together, these drive glioma aggressiveness in a tension-dependent feedback loop. Unprocessed blot: Supplementary Fig. 9.