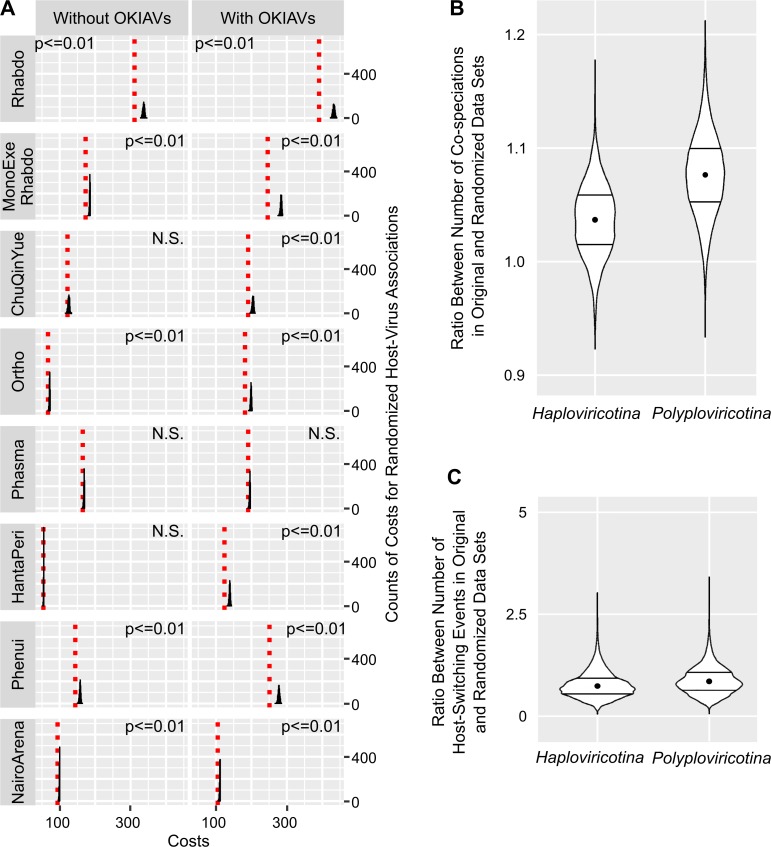
Fig 5. Analysis of host-virus phylogenetic co-segregation.
A: Histograms of costs for co-segregation of randomized host associations (1000 iterations) compared to the original host associations (red dotted lines) for all phylogenies, without (left) and with (right) OKIAV sequence inclusion, calculated with Jane [17]. The p-values of all z-tests indicate an increase in costs of over 5% above the original costs. N.S. indicates non-significant cost differences. B: Ratio between number of co-speciations in original and randomized data sets calculated for Haploviricotina and Polyploviricotina with CoRe-PA [18]. C: Ratio between number of host switching events in original and randomized data sets calculated for Haploviricotina and Polyploviricotina with CoRe-PA.