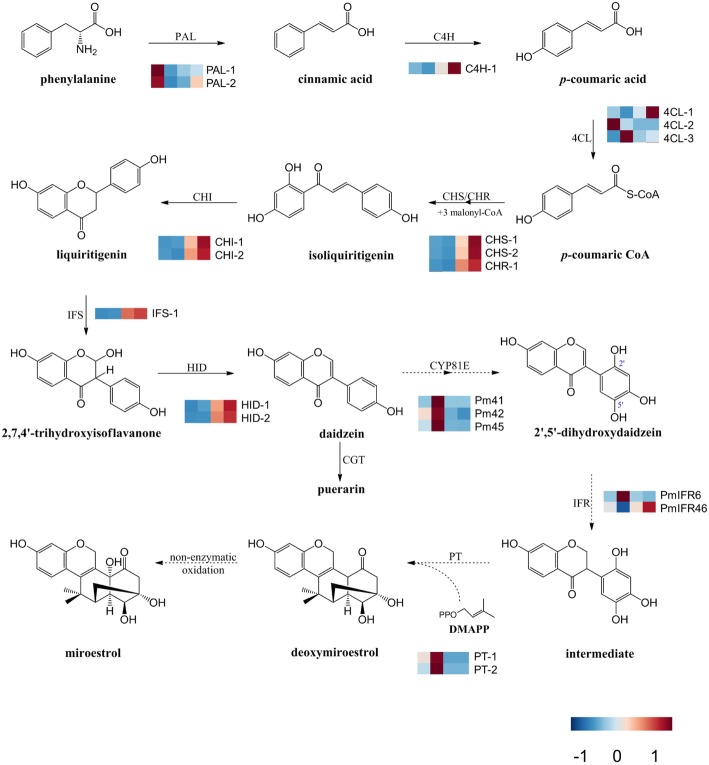
Fig. 1.
Proposed miroestrol biosynthetic pathway in P. mirifica. The heatmap following each gene name indicates the relative differential expression of genes in young leaves, mature leaves, tubers without cortices, and cortices of P. mirifica, respectively. Enzyme abbreviation: PAL; Phenylalanine ammonia-lyase, C4H; Cinnamate 4-monooxygenase, 4CL; Coumarate-CoA ligase, CHS; Chalcone synthase, CHI; Chalcone isomerase, IFS; 2-hydroxyisoflavone synthase, HID; 2-hydroxyisoflavone dehydratase, CGT; C-glycosyltransferase, CYP81E; Cytochrome P450 subfamily 81E, IFR; Isoflavone reductase, PT; Prenyltransferase. Although the assembled unigenes are hypothetical candidate genes, a comparative transcriptome analysis using P. lobata is a promising way to study and identify species-specific and evolutionary conserved pathways involved in the highly complex biosynthesis of miroestrol