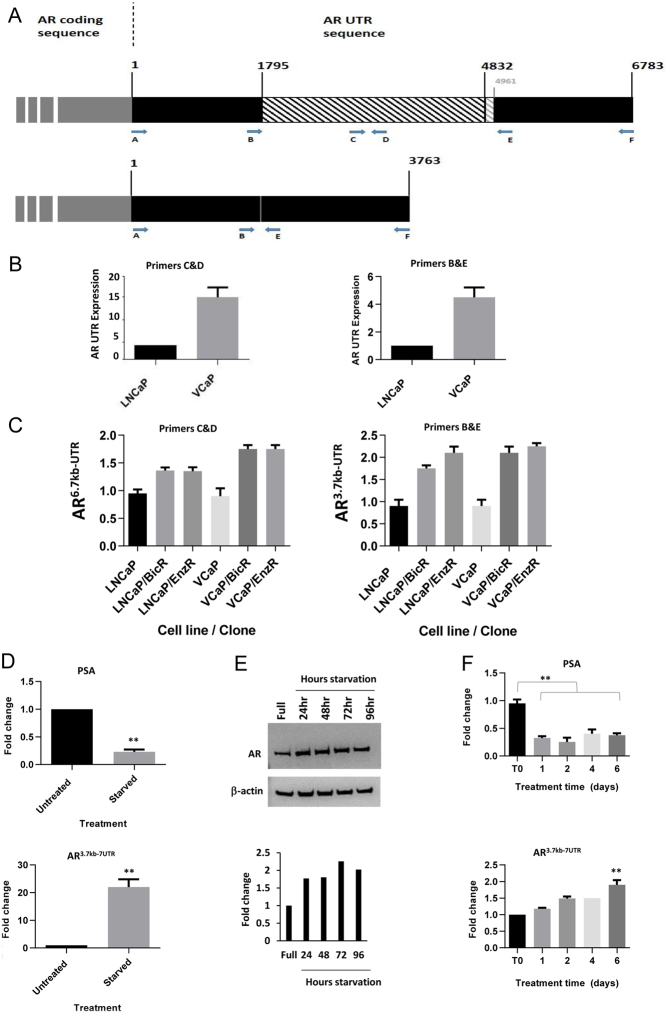
Figure 3.
AR 3′UTR variant increases due to hormonal starvation or AR-antagonists treatment. (A) Schematic diagram indicating the binding location of qPCR primer sets used for detecting either a portion of the 6.7 kb 3′UTR of AR (upper panel) or the novel shorter AR 3 kb transcript (lower panel). Numbers represent base locations in the 3′UTR. (B) qPCR analysis of the AR UTR utilising the indicated primers sets in parental LNCaP or VCaP cells. (C) qPCR analysis of AR6.7kb-UTR variant and AR3.7kb-UTR variant in parental and resistant LNCaP and VCaP clones. (D) qPCR analysis of PSA (upper panel) and AR3.7kb-UTR (lower panel) in LNCaP cells grown in starvation medium for 72 h. (E) Western blot for AR protein levels in hormonally starved LNCaP cells for 0–96 h. Normalised to β-actin, with associated densitometry underneath. (F) qPCR analysis of PSA (upper panel) and AR3.7kb-UTR variant (lower panel) in LNCaP cells treated with bicalutamide (10 µM) for 6 days. Data represent the mean and s.e. from three independent replicates. Data are normalised to housekeeping genes GAPDH, RPL19 and β-actin. Statistical analysis * (P = 0.05), ** (P = 0.01) from a t-test.

 This work is licensed under a
This work is licensed under a