Figure 1.
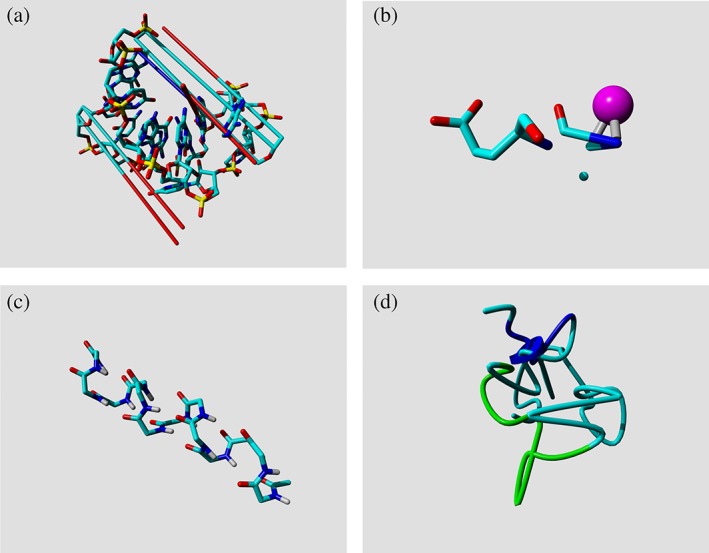
Examples that explain why not all PDB entries are equally useful for PSB studies. (a) The long lines indicate very long bonds in 1I4C. These seem to be caused by incorrect formatting of the PDB entry. Coordinates are written with only two characters before the decimal point, which makes them lose the minus sign when a coordinate is below −9.999; for example, an X‐coordinate like −11.236 becomes 11.236. (b) The N‐terminus of the azurin structure, 1AG0, once consisted of two half alanines with a copper ion in the middle. This problem has recently been solved, but the PDB provides no easy mechanism for correction tracking. (c) This small polyglycine helix (1CEK) is actually the result of a complex biophysical experiment. There is nothing wrong with this structure, but we believe that it should not be used in PDB‐wide computational studies. (d) Something went very wrong when solving this protein structure (2PDE, subunit‐binding domain of dihydrolipoamide acetyltransferase), the very short helix that is indicated by a short blue cylinder works as a chaotic attractor through which the chain passes seven times
