Figure 4.
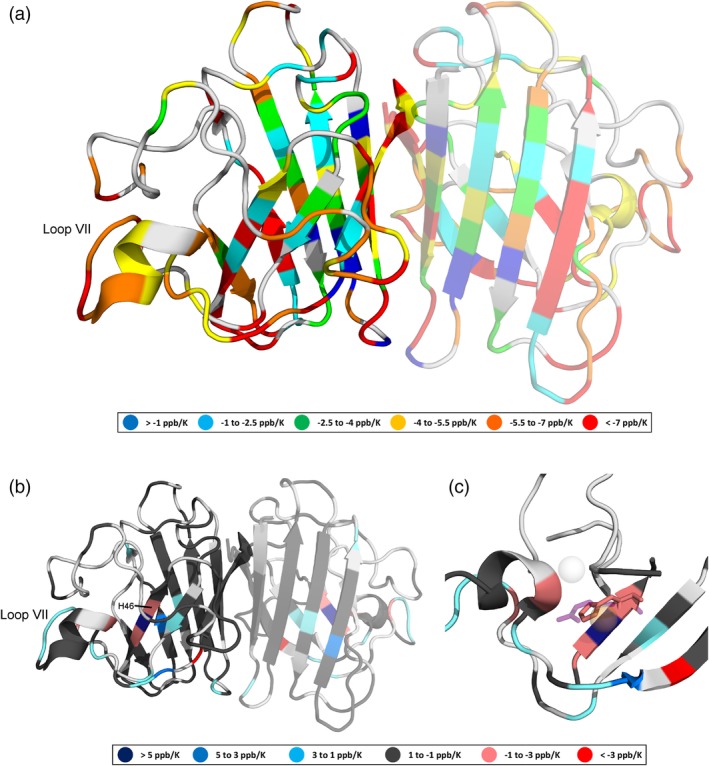
(a) Amide proton temperature coefficients (determined using chemical shifts referenced to DSS) for pWT apoSOD12SH, (b) temperature coefficient differences (relative to pWT) resulting from the H46R mutation, and (c) the metal‐binding region of apoSOD12SH showing overlaid H46 (pink) and R46 (purple, semitransparent) side chains. Light gray indicates that data are not available. The monomeric, rather than dimeric form of apoSOD12SH predominates, but in (a) and (b), we project the data onto a dimeric SOD1 structure (PDB ID: 2AF2)26 with a semitransparent second subunit in order to illustrate the residues that form the dimer interface and present two views (one rotated ∼ 180° relative to the other). In holoSOD1 (PDB ID: 1HL5),27 the H46 residue coordinates the Cu2+ ion (orange, semitransparent), while in H46R (PDB ID: 1OZT) the positively charged guanidinium group of the R46 side chain sits between the vacant Cu2+ and Zn2+ (light gray, semitransparent) binding sites. Figure prepared using PyMOL28
