Figure 5.
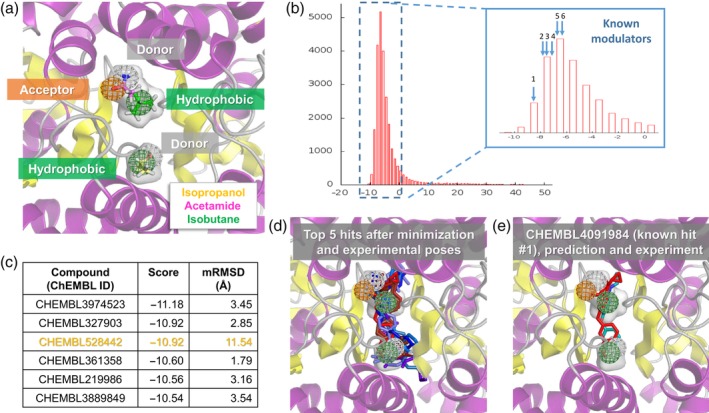
Compounds obtained from virtual screening of our PM against the ChEMBL 25 library of small molecules. (a) PM constructed based on the snapshot displayed in Figure 4c, using the binding position and orientation of three probe molecules: acetamide, isobutane, and isopropanol. C atoms are colored by probe as in previous figures, H, N, and O atoms are colored white, blue, and red, respectively. Hydrogen bond donor (gray) and acceptor (orange) features were used for acetamide (both donor and acceptor) and isopropanol (donor), and hydrophobic features (green) were used for isopropanol and isobutane. Selected probes span the gray transparent volume, which is displayed in other panels too for comparison. (b) Histogram showing AutoDock Vina scores from querying the ChEMBL 25 database with Pharmit. The majority of scores are negative (corresponding to favorable binding energies). The inset enlarges this region with negative scores and indicates the scores for six known AMPAR modulators listed in ChEMBL 25 (see Table 1). (c) AutoDock Vina score and minimization RMSD (mRMSD) between docked compounds and the input pharmacophore features are listed for the top hits (compounds with the highest binding affinity or most negative scores).One of the compounds (highlighted in orange) has a high mRMSD value, meaning its preferred binding pose after minimization is not consistent with the input PM. (d). Optimized binding poses of the top five hits shown in various shades of blue, along with the experimentally resolved modulators (R,R)‐2a, (R,R)‐2b, and 11m from PDB structures 3BBR, 4U5B, and 5OEW in red. (e) The predicted binding pose of the top known modulator (11m) and its experimental pose show good agreement with the PM. See also Figure S3 for additional hits and experimentally determined compounds (Table 1) that match the PM. PDB, protein data bank; PM, pharmacophore model
