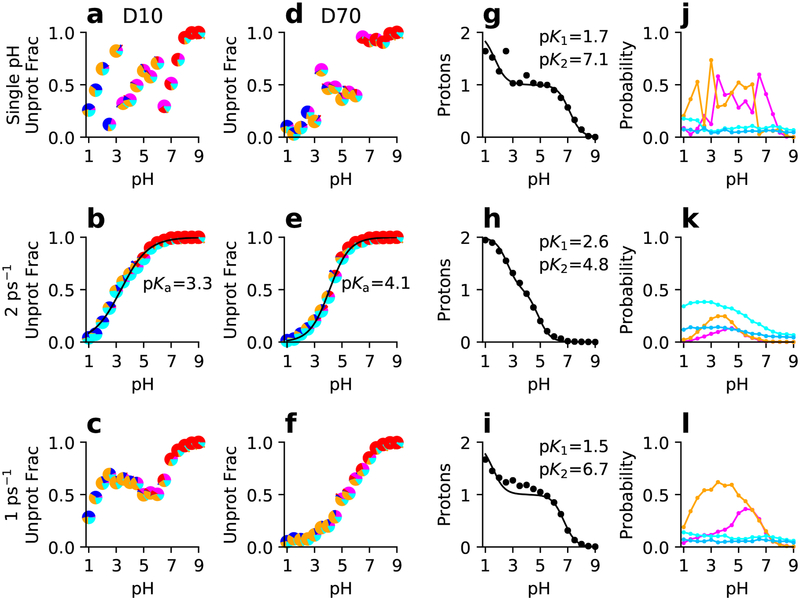
Figure 6: Simulated titration of the linked residues D10 and D70 in RNase H from single-pH and replica-exchange simulations.
From top to bottom rows, single-pH and replica-exchange simulations with different exchange attempt frequencies are shown. (a-f) Unprotonated fraction as a function of pH and best fit to the residue-specific HH equation for D10 (a-c) and D70 (d-f). Each data point is a pie chart showing the probability of the four protonation states: D10H/D70H (blue), D10−/D70− (red), D10H/D70− (magenta), and D10(−)/D70H (orange). Cyan indicates that one of the residues is in a mixed state. (g-i) Total number of protons as a function of pH and best fit to the linked-titration model. (j-l) Probabilities of the singly-protonated (magenta for D10H/D70− and orange for D10−/D70H) and mixed states (light cyan for D10 titration and dark cyan for D70 titration) as functions of pH. The experimental macroscopic pKa’s are 6.1 and 2.6 for D10 and D70, respectively.