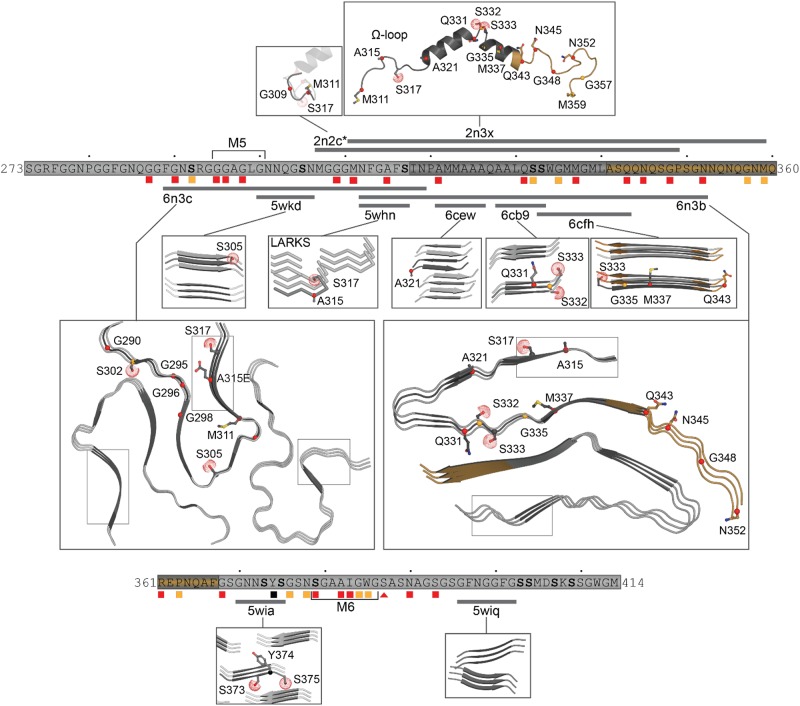
FIGURE 5.
Structures of the C-terminal domain. Structures shown with mutation sites as sticks and phosphorylation sites with dots at the SG atom. Simplified primary sequence annotations as in Figure 1. Structure 2n2c (Lim et al., 2016) is highly similar to that of 2n3x (Jiang et al., 2016), thus only a portion is shown here, highlighting the non-overlapping residues M307-G310. Representatives of the short segments that form steric zippers or LARKS (5wkd, 5whn, 6cew, 6cb9, 6cfh, 5wia, 5wiq) (Guenther et al., 2018a) are shown in darker gray with neighboring strands as determined by symmetry in lighter gray. The R-shaped polymorph (6n3c) (Cao et al., 2019) with chain A in darker gray and only three filaments and immediate adjacent layers shown for clarity. A representative dagger-shaped fibril-forming polymorph (6n3a) (Cao et al., 2019) is shown with chain B in darker gray with two filaments and immediate adjacent layers. Note different shapes of LARKS segments within 6n3c and 6n3b (thin boxes). For clarity, mutation or phosphorylation sites shown only on the primary strand in all images.