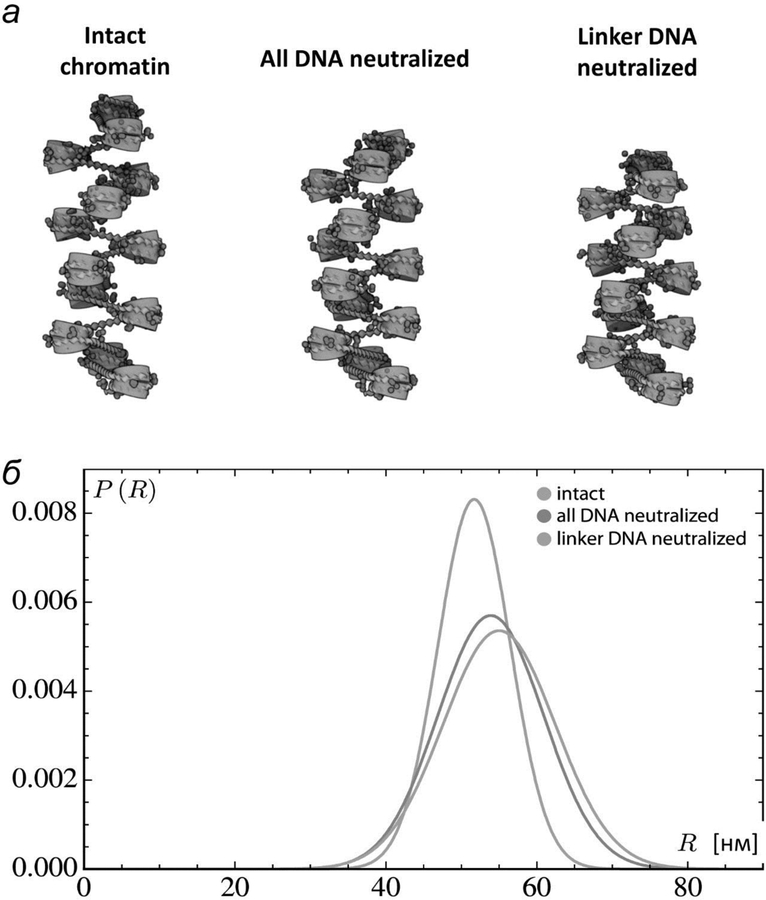
Fig. 5.
Computational simulation of the chromatin fiber: distant internucleosomal interaction are increased after DNA charge neutralization. а – Models of chromosomal fragments based on the average configurations of the central residues of simulated structures with 13 intact nucleosomes on fully charged, fully neutralized, and linker-neutralized DNA The central residues are expected to be representative of the configurational states adopted by a long fiber with evenly spaced nucleosomes. б – Distribution of distances between the first and last base pairs on simulated chromatin constructs with 13 intact nucleosomes, spaced at 177-bp increments (30-bp linkers), on fully charged, fully neutralized, and linker-neutralized DNA.