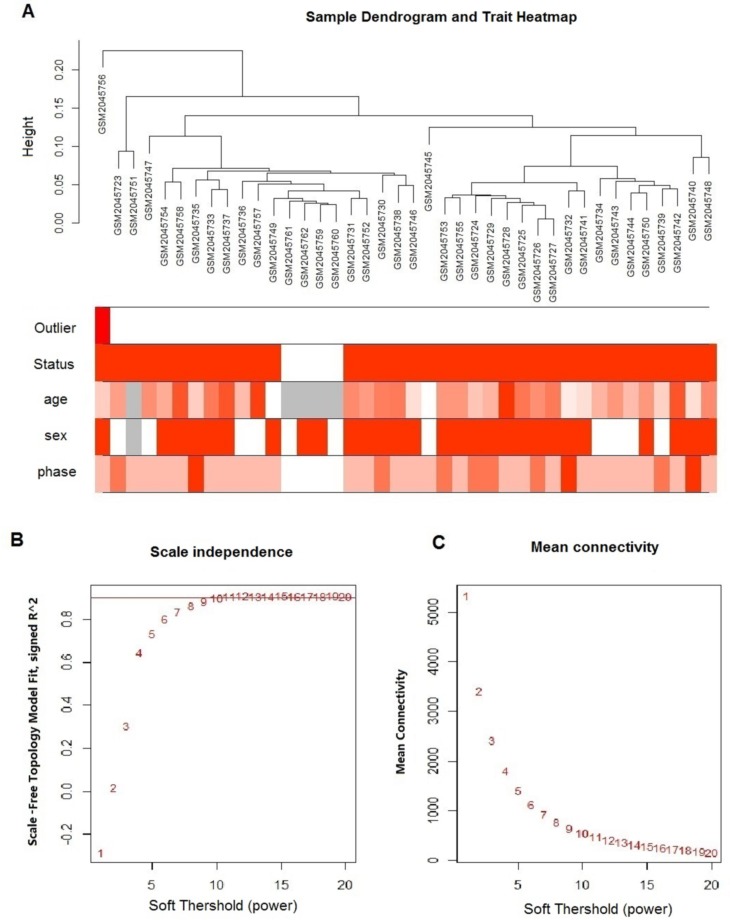
Figure 1.
Clustering of samples and determination of soft-thresholding power. (A) The clustering heat map was based on expression data of GSE77191, including 26 CML CP, 7 CML AP, 3 CML BP, and 4 normal samples. The top 10,000 genes with the highest SD values were used for WGCNA analysis. Color intensity was proportional to sample outliers, disease status (CML and normal control), phase (normal control, CP, AP, BP), sex, and age. (B) Analysis of the scale-free fit index (y-axis) for various soft-thresholding powers (β value of x-axis). (C) Analysis of the mean connectivity (y-axis) for various soft-thresholding powers (β value of x-axis).