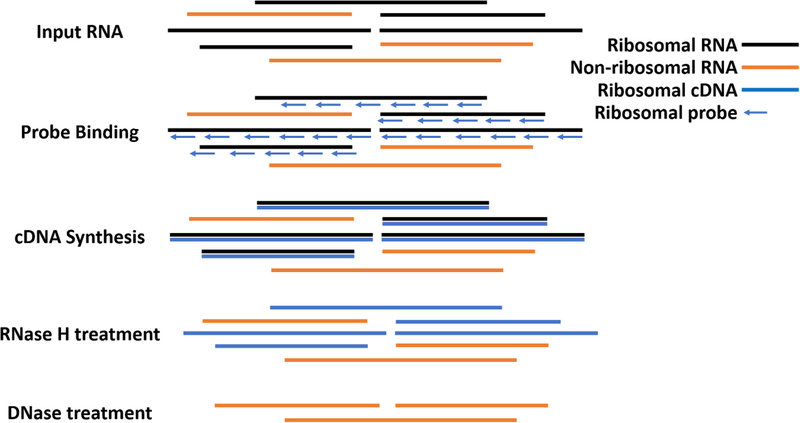
Fig. 1. Workflow for reverse-transcriptase mediated ribosomal depletion from total RNA.
To perform ribosomal RNA (rRNA) depletion, total RNA is first extracted, DNase treated and subsequently purified with RNAClean XP Beads (Agencourt). DNA-free RNA is then bound to oligonucleotide probes designed to bind to rRNA from mosquito species in Aedes, Culex and Anopheles genera that are in the reverse complement orientation to both the long and short ribosomal subunit and 12S and 16S mitochondrial rRNA. The RNA with bound oligos is then subjected to reverse transcription using Avian Myeloblastosis Virus (AMV) Reverse Transcriptase (NEB). RNA that is reverse transcribed to cDNA is then digested using RNase H, which selectively destroys RNA in an RNA:DNA hybrid. Remaining DNA is then digested using DNase I (NEB), leaving mostly non-ribosomal RNA which is then used for library preparation.