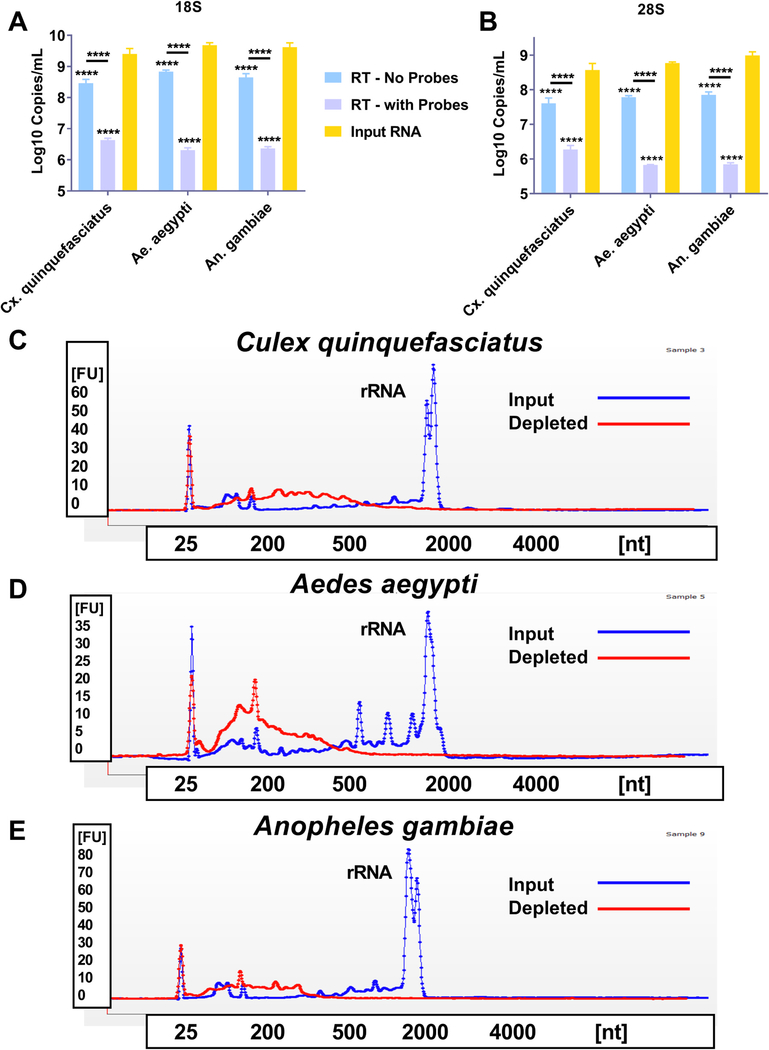
Fig. 3. Reverse Transcriptase mediated ribosomal RNA (rRNA) depletion is effective against mosquitoes from three distinct medically relevant genera.
Total RNA was extracted from three distinct pools of whole mosquitoes from three medically relevant genera; Culex (Cx.) quinquefasciatus, Aedes (Ae.) aegypti and Anopheles (An.) gambiae. The RNA was treated with DNase I and then purified; this will now be called input RNA. An aliquot was then taken, and reverse transcribed to cDNA using AMV reverse transcriptase (RT) and DNA probes specific for mosquito ribosomal RNA (RT – with Probes) or in the absence of probes (RT – no probes). The samples were then treated with RNase H and DNase I to remove the RNA present in an RNA:DNA hybrid and cDNA, respectively. The samples were then purified and subjected to qRT-PCR with primer probe combinations specific for 18S or 28S rRNA (A and B). The input RNA and RT – with Probes were then assessed using a Bioanalyzer. Panels C-E show a representative trace for each of the three mosquito species tested, Cx. quinquefasciatus (C), Ae. aegypti (D), An. gambiae (E). The blue trace for each panel shows the input RNA and the red trace shows the RT – with Probes treated RNA. The peak present at roughly 40 s in each trace is the peak for both 18S and 28S rRNA. All statistical tests were performed by Two-Way ANOVA with Tukey test for multiple comparisons. ****Indicates p-value < 0.0001. All stars without an accompanying bar are statistical comparisons between the input RNA and the other group. The other comparisons with a bar are statistical comparisons between the two groups that are below the corresponding bar.