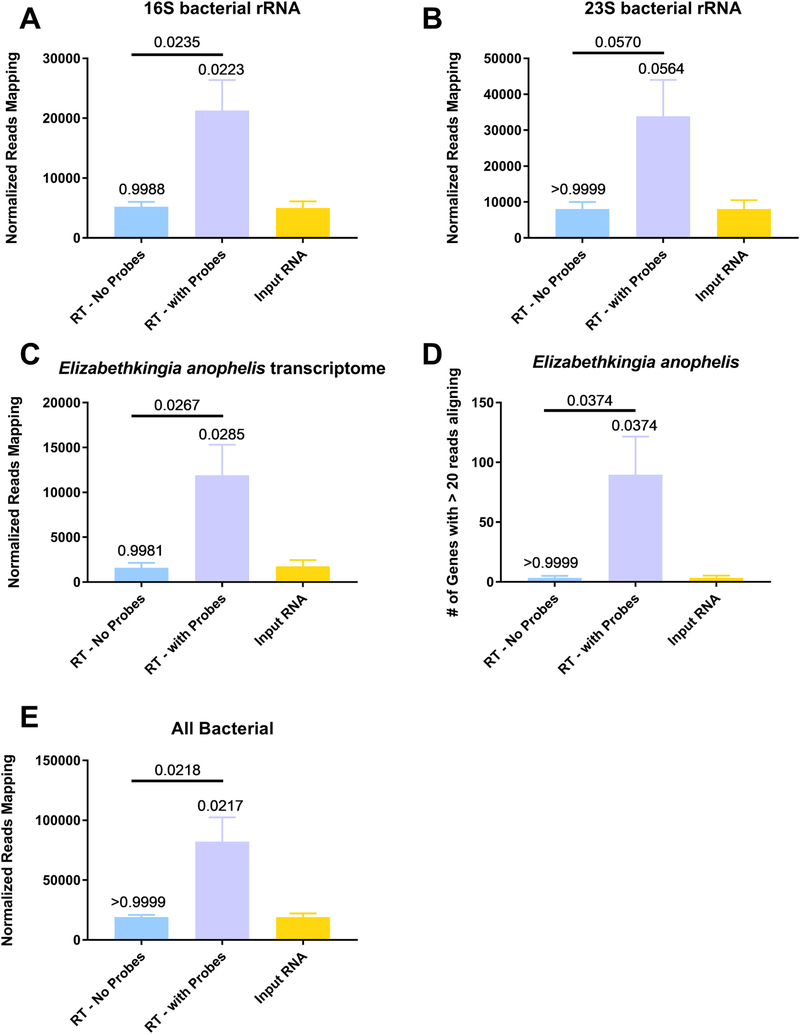
Fig. 6. Percent of reads to bacterial rRNA and host transcripts are increased in depleted samples.
Anopheles gambiae mosquitoes were exposed to an infectious bloodmeal containing 107 PFU of West Nile virus strain NY99. The following day, midguts were dissected and the residual bloodmeal was spread onto a CloneSaver FTA card (GE Healthcare, USA) and then soaked in RNAlater solution to stabilize the nucleic acid and facilitate dispersion. Total nucleic acid was then extracted, and DNase treated. This is considered the input RNA. DNase-free RNA was then reverse transcribed using either ribosomal RNA specific probes (RT – with Probes) or without probes (RT – no Probes). The samples were then treated with RNAse H and DNase I and purified. The samples were then subjected to library preparation and sequenced on an Illumina MiSeq. Reads were then demultiplexed and subsequently trimmed using BBDuk. Each sample was then normalized to contain 1.5 million reads to allow for direct comparisons. Duplicate reads were removed using Clumpify and then unique reads were mapped using either BBSplit to the appropriate reference sequence, 16S bacterial rRNA (A), 23S bacterial rRNA (B), Elizabethkingia anophelis transcriptome (C and D) and the NCBI bacterial genome database (E). The reads called for each gene was determined using BBMap with output flag rpkm (D). All statistical tests were performed by One-Way ANOVA with Tukey test for multiple comparisons. The p-values are indicated as numbers. All p-values without an accompanying bar are statistical comparisons between the input RNA and the other group. The other comparisons with a bar are statistical comparisons between the two groups that are below the corresponding bar.