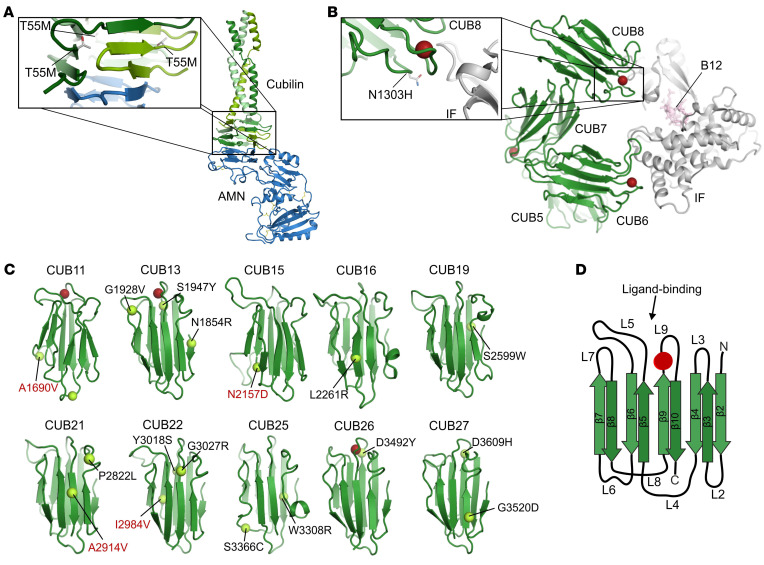
Figure 3. Structural modeling of CUBN missense variants.
(A) Location of the cubilin T55M missense variant in RCSB entry 6GJE. The variant is located in the hydrophobic core of the cubilin β-helix that interacts with amnionless. (B) Location of the cubilin N1303H missense variant in RCSB entry 3KQ4. The variant is located in CUB domain 8, close to the interface with IF/B12. Red spheres represent Ca2+ ions binding to the CUB domains. (C) Location of the cubilin missense plus the 4 GWAS variants (in red) in in silico structural models of CUB domains 11–27. Red spheres represent Ca2+ ions predicted to bind CUB domains 11, 13, and 26. The variants S1947Y and D3492Y are notably close to these Ca2+-binding motifs. (D) Example of 1 CUB domain with β-sheets (β2–10) and loops (L2–9).