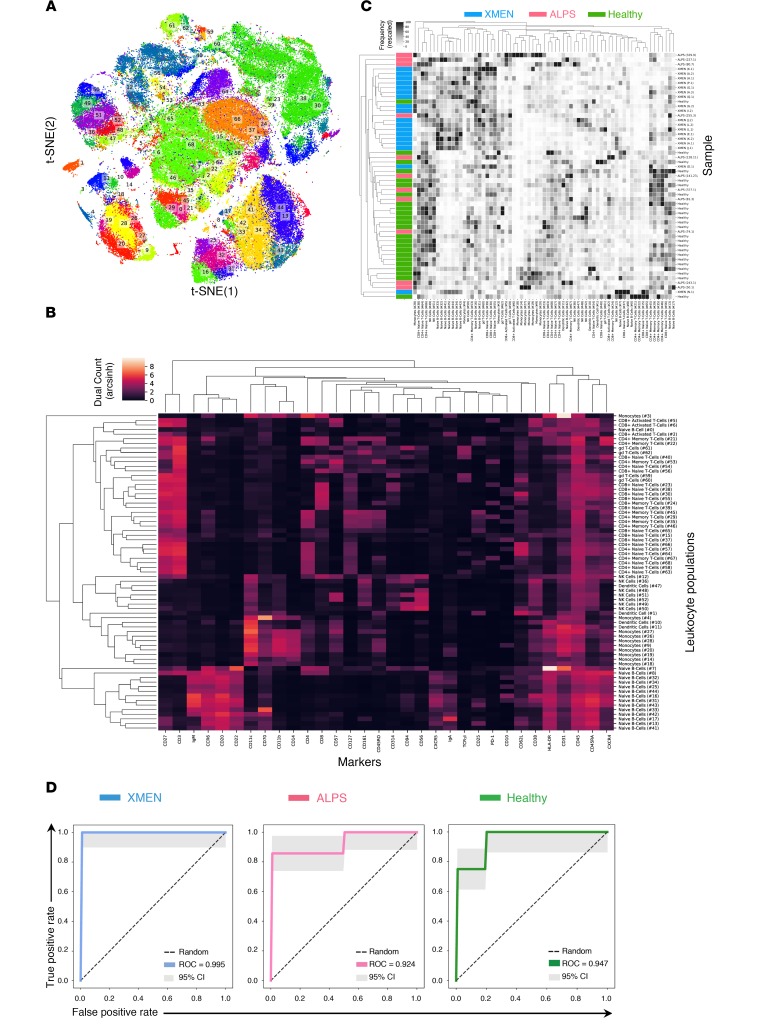
Figure 4. Deep immunophenotyping of PBMCs shows distinctive immune subsets for XMEN compared with HCs and ALPS.
HAL-x on CyTOF data acquired from PBMCs from patients with XMEN (n = 18), patients with ALPS (n = 11), and HCs (n = 24) identified 69 CoD. (A) 2D projection of the identified CoD as visualized by t-distributed stochastic neighbor embedding (t-SNE). (B) Dendrogram showing these CoD based on their abundance of surface epitopes. (C) Dendrogram of the frequencies of these CoD based on unsupervised grouping showing the clustering of patients with XMEN (blue), patients with ALPS (magenta), and HCs (green). (D) ROCs for the random forest classification of XMEN, ALPS, and HCs based on the frequency of their CoD. The AUC value is the mean AUC taken after a 4-fold cross-validation. Each fold of the cross-validation represents a different separation (of the patients) into training and testing sets. 95% CIs are shown (gray).