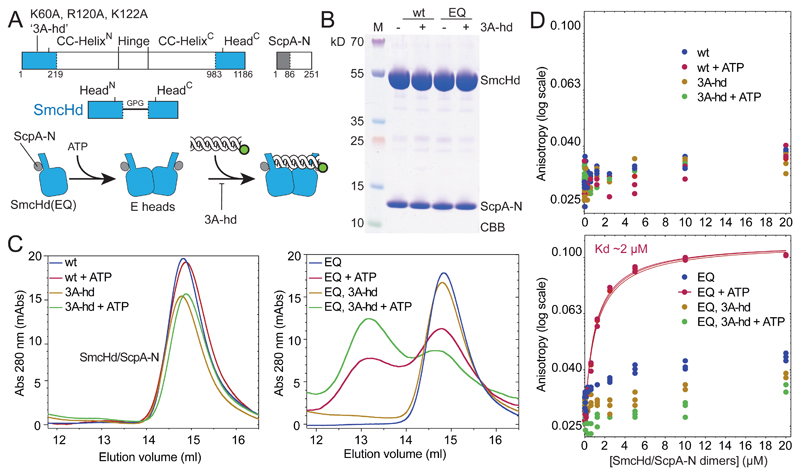
Figure 4. ATP dependent head/DNA association.
(A) Construction of SmcHd/ScpA-N with and without the ‘3A-hd’ mutations (K60A, R120A, K122A). The amino-terminal His6-tag on ScpA-N is omitted from the scheme for simplicity.
(B) CBB staining of purified preparations of SmcHd/ScpA-N constructs with and without EQ and 3A-hd mutations.
(C) Gel filtration profiles of SmcHd/ScpA-N preparations eluted from a Superdex 200 10/300 column. Samples were eluted in buffer supplemented with or without 1 mM ATP. Elution curves for proteins without and with the EQ mutation are shown in the left and right panels, respectively.
(D) DNA binding measurements using fluorescence anisotropy titrations as in Figure 2C. Data points for proteins without and with the EQ mutation are displayed in the top and bottom panels, respectively. Data points and fits for three experiments are shown. The estimated Kd for head/DNA association is calculated under the assumption of complete dimerization of SmcHd(EQ)/ScpA-N. The actual Kd may thus be somewhat lower.
See also Figure S4 and Table S3.