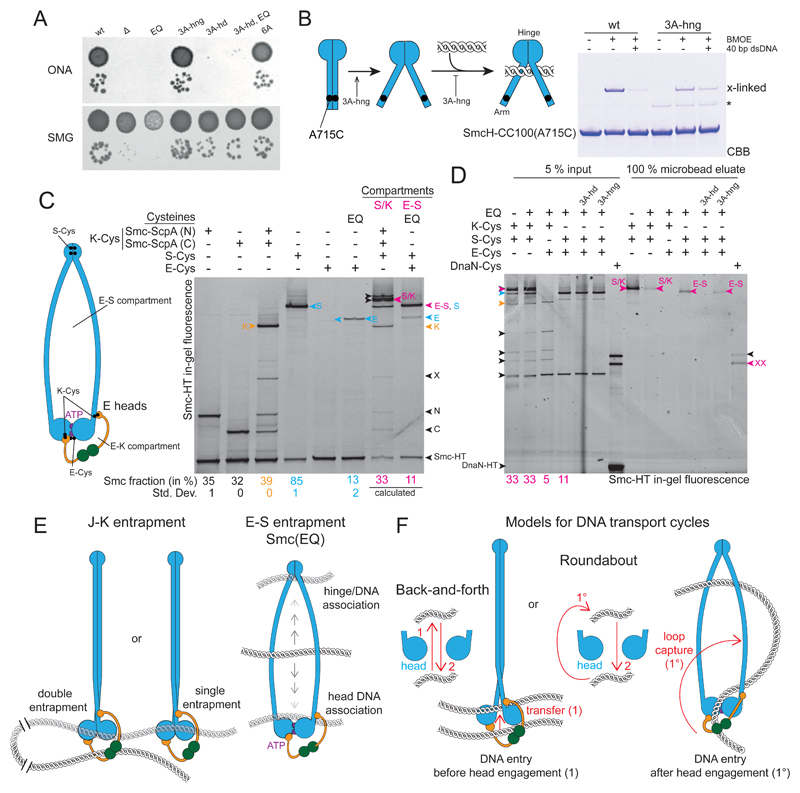
Figure 7. Hinge and head mutations promote S compartment opening and DNA entrapment.
(A) Dilution (81-fold and 59000-fold) spotting of 3A-hng, 3A-hd and 6A mutants. As in Figure 2D. A corresponding ONA image after 12 h incubation is shown in Figure S7A.
(B) Cysteine cross-linking of purified SmcH-CC100(A715C) protein in presence and absence of 10 μM 40 bp dsDNA. The asterisk marks an impurity; see also Figure 2B.
(C) Identification and quantification of cross-linked species of E heads Smc/ScpAB. using cysteine pairs in isolation and combination. As in Figure 1B. Circular species are indicated in magenta colors and denoted as ‘S/K’ for the SMC/kleisin ring and ‘E-S’ for the E-S compartment. Species with E heads cross-linked only (‘E’) are labelled in blue colors. All other labeling as in Figure 1B. Note that species S and E-S co-migrate in the gel. Mean fraction (and standard deviation) of species E in Smc(EQ) was quantified from three replicate experiments. Quantification of all other species as in Figure 1B (Table S2). The fraction of E-S species was calculated under the assumption of normal hinge dimerization in E heads Smc(EQ)/ScpAB (Bürmann et al., 2017).
(D) DNA entrapment in the E-S compartment of Smc(EQ)/ScpAB. Chromosome entrapment assay with agarose microbeads. As in Figure 1D. Arrowheads in magenta color denote circular protein species, S/K for the SMC/kleisin ring and E-S for the E-S compartment.
(E) Summary of main findings.
(F) Models for DNA transport cycles: DNA entry into the S compartment by passage between separated Smc heads is shown on the left-hand side (‘Back-and-forth’). DNA entry into the S compartment during head engagement by loop capture displayed on the right-hand side (‘Roundabout’).
See also Figure S7 and Table S2.