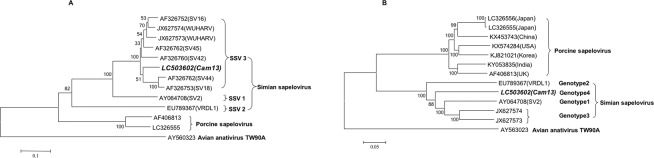
Figure 3.
Phylogenetic relationships among SSV strains. The amino acid and nucleotide sequence alignment was performed using ClustalW software, with an Avian anativirus TW90A as the out-group. The genetic distance was calculated by Kimura’s two-parameter method. Phylogenetic trees with 1,000 bootstrap replicates were generated by the Neighbor-Joining method using MEGA (ver. 6) based on the aa sequence of VP1 (A) and the entire genome (B) of SSV, PSV, and TW90A. Scale bar indicates amino acid or nucleotide substitutions per site. Strain Cam13 strain is shown in bold italic letters. The genotype is tentatively named based on the findings of our present study.