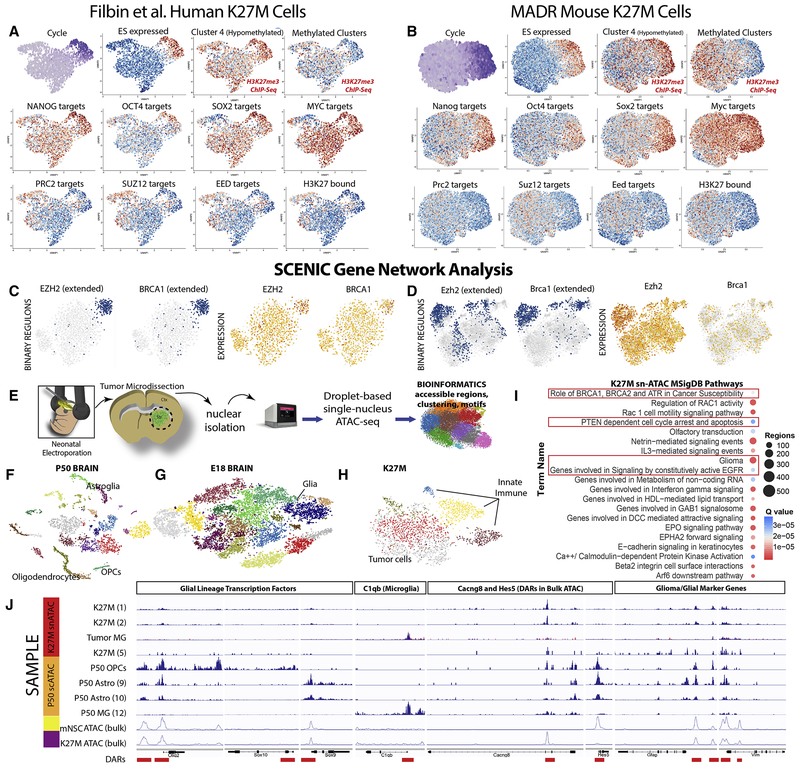
Figure 7: H3.3 K27M Transcriptional Network and snATAC-seq Analysis.
(A-B) t-SNE featureplots depicting cell type-specific upregulation NANOG, OCT4, SOX2, MYC target genes, and embryonic stem cell (ES)-associated gene sets and the underexpression of PRC2, SUZ12, EED, and H3K27-bound gene sets for human cells (A) and analogous genes/genesets in mouse (B).
C-D) Binary regulon activity and corresponding scRNA-seq featureplot for EZH2, BRCA1 (C; Filbin et al. dataset), Ezh2, and Brca1(D; K27M mouse dataset).
E) Schematic of snATAC-seq sample preparation
F) tSNE of sc- and snATAC datasets from P50, F) E18) and G) K27M mouse brains
I) K27M snATAC-identified MSigDB pathways
J) Genome browser alignments of snATAC-seq, scATAC-seq, and bulk ATAC-seq. *-Tumor MG is an overlaid(red/black) alignment of snATAC-seq microglial clusters captured with the K27M cells.