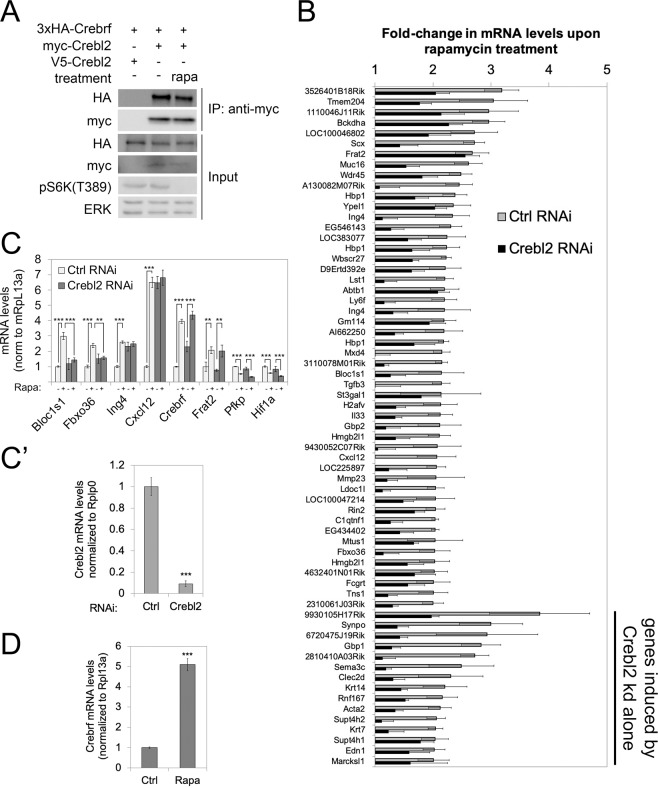
Figure 1.
Rapamycin induced transcription is partially dependent on Crebl2 in MEFs. (A) Crebl2 and Crebrf bind each other. Co-IP of 3xHA-CREBRF with myc-CREBL2 in HEK293T cells. V5-Crebl2 is used as a negative control for the myc-IP. (B) Crebl2 is required for induction of gene expression in response to rapamycin (20 nM, 12 h). Fold change after rapamycin treatment from Illumina BeadChip analysis for the top inducing genes in control (grey) and Crebl2 knockdown (black) TSC2−/− MEFs. Error bars represent biological triplicates except Luciferase control knockdown samples which were duplicates. (C,C’) Knockdown of Crebl2 prevents rapamycin (20 nM, 12 h) induction of target genes in TSC2−/− MEFs. mRNA levels by qPCR, normalized to Rpl13a. Frat2 is shown as unaffected control and Pfkp, Hif1a are shown as rapamycin-repressed controls. Knockdown efficiency of Crebl2 shown in (C’). Error bars represent technical triplicates. ***p < 0.001, **p < 0.01 by two-way ANOVA. (D) Crebrf is transcriptionally induced by rapamycin treatment (20 nM, 12 h) in TSC2−/− Mefs. mRNA levels by qRT-PCR, normalized to Rpl13a. Error bars represent technical triplicates. ***p < 0.001 by t-test. All panels: error bars = SD.