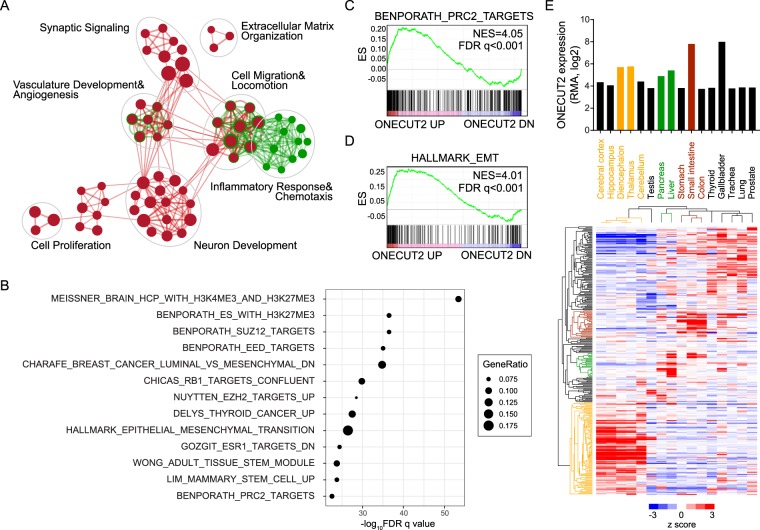
Figure 4.
ONECUT2 overexpression activates lineage-specific genes and oncogenic pathways. (A) A total of 1285 DEGs were subjected to DAVID GO analysis. An enrichment map was constructed by using Cytoscape with the Enrichment Map plugin. Red nodes represent enriched GO biological pathways of upregulated genes, and green nodes and rings represent enriched GO biological pathways of downregulated genes (p < 1e-05, FDR q < 1e-04, overlap cutoff > 0.68). Nodes and ring sizes are proportional to the total number of genes in each pathway. Edge thickness represents the number of overlapping genes between nodes. GO pathways of similar functions are clustered together and are marked with circles and labels. (B) Top 10 gene sets in MSigDB that overlapped with genes upregulated by ONECUT2 overexpression. (C,D) GSEA of PRC2 targets (C) and EMT(D) gene signature on ONECUT2 DEGs. Genes were ordered by expression fold changes by ONECUT2 overexpression in descending order from left to right. (E) Lineage-restricted expression pattern of ONECUT2-upregulated genes. Hierarchy clustering of expression of ONECUT2-upregulated genes across multiple normal tissues, alongside with expression of ONECUT2.