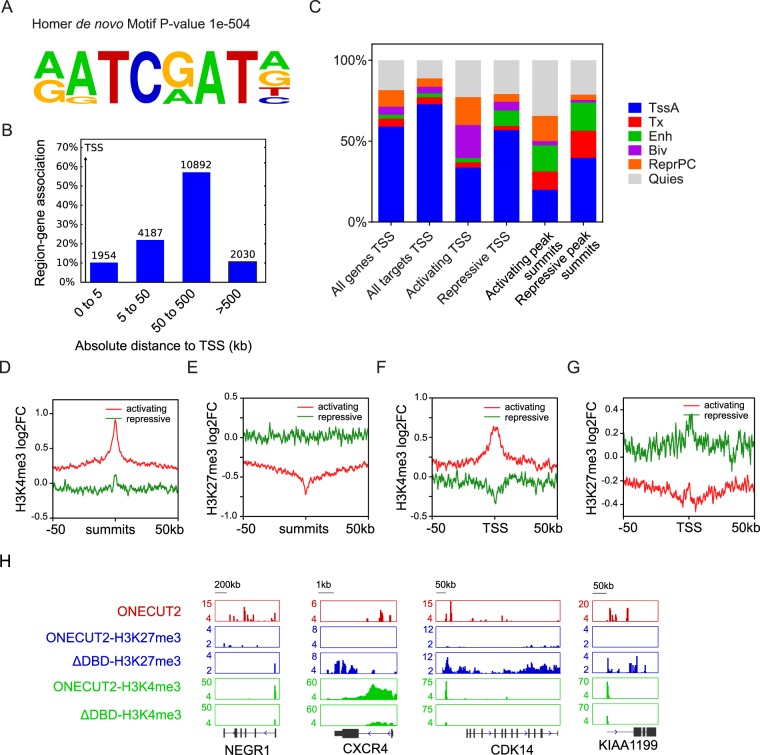
Figure 5.
ONECUT2 binds and modulates the activity of bivalent chromatin. (A) Homer motif analysis of high confidence ONECUT2 peaks. (B) GREAT analysis of high confidence ONECUT2 peaks shows distribution of gene-region assignments around TSS. (C) Percentage plot of Roadmap A549 cell chromatin states around activated and repressed ONECUT2 target gene TSS, as well as summits of activating and repressive peaks. Chromatin state around the TSSs of all protein-coding genes and all ONECUT2 target genes are shown as control. (D,E) Plot of average H3K4me3 (D) and H3K27me3 (E) log2 fold change by ChIP-Seq (A549-ONECUT2 versus ΔDBD) around activating and repressive ONECUT2 peaks. (F,G) Plot of average H3K4me3 (F) and H3K27me3 (G) log2 fold change by ChIP-Seq (A549-ONECUT2 versus ΔDBD) around TSS of genes associated with activating and repressive ONECUT2 peaks. (K) Representative illustration of H3K4me3 and H3K27me3 signals around four selected loci alongside ONECUT2 binding signals in A549-ONECUT2 cells.