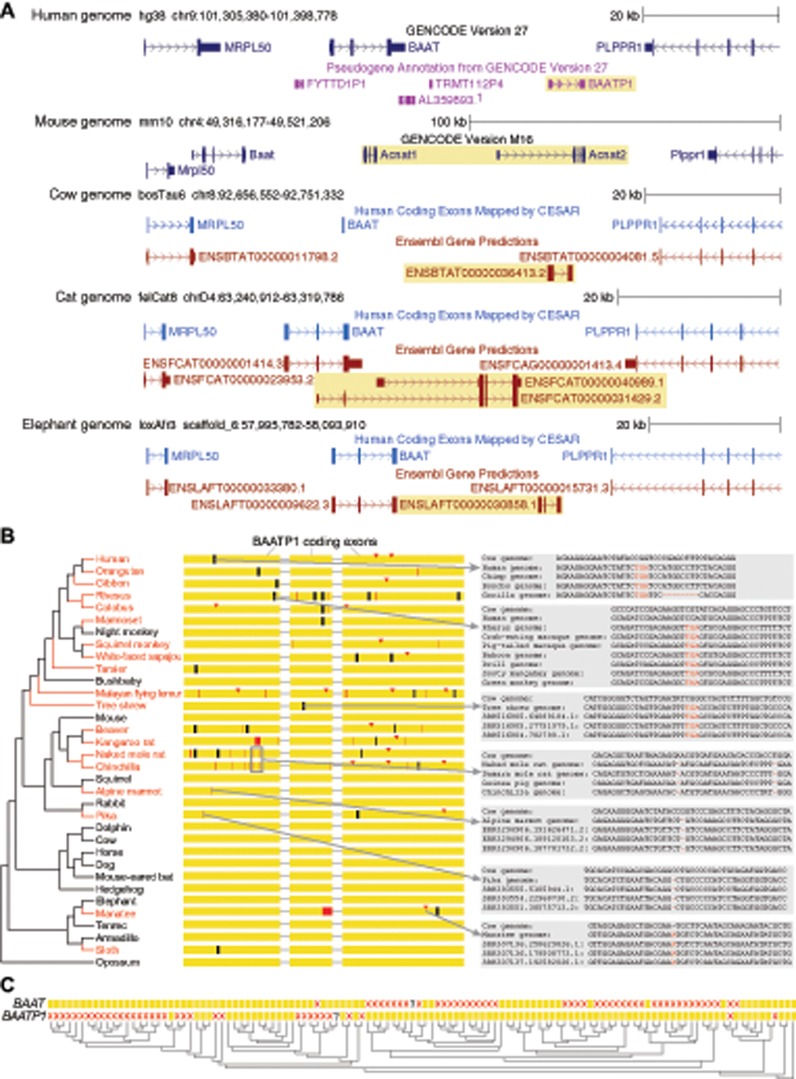
Fig. 2.
—Evolution of a second BAAT gene (BAATP1) that arose by a tandem duplication. (A) UCSC genome browser (Haeussler et al. 2019) screenshot of the human locus containing BAAT and the surrounding genes (MRPL50 and PLPPR1) shows the presence of an intron-containing pseudogene BAATP1 (yellow background) downstream of BAAT, suggesting that this pseudogene copy arose by a tandem duplication. This pseudogene aligns to an intact gene in other placental mammals, as shown by the Gencode or Ensembl gene annotation (Cunningham et al. 2019). In the mouse, a tandem duplication of the human BAATP1 ortholog occurred, giving rise to two genes annotated as Acnat1 and Acnat2. In addition to Ensembl, we show human genes that are mapped by CESAR to the orthologous cow, cat, and elephant locus via the genome alignment (Sharma et al. 2016; Sharma and Hiller 2017). As CESAR only projects intact human genes, it cannot annotate intact BAATP1 orthologs (yellow background) in other species. In cow, CESAR only annotates the intact second exon of BAAT, as the other exons have inactivating mutations (fig. 1). (B) Similar to BAAT, BAATP1 is lost repeatedly in placental mammals (visualization as in fig. 1). (C) Visualizing the presence (yellow box) or loss (red cross) of BAAT and BAATP1 in 118 mammals shows that losses of these genes mostly occurred in a reciprocal fashion. A question mark indicates that it is uncertain whether the gene is present or lost.