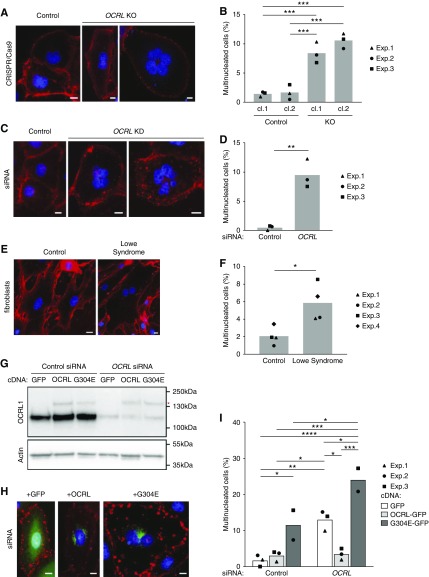
Figure 2.
Cells lacking OCRL exhibit increased multinucleation. Control and OCRL-depleted cells were stained with fluorescently conjugated wheat germ agglutinin (red) and DAPI (blue) to visualize cell borders and nuclei, respectively. (A) Representative images of mononuclear cells in control CRISPR/Cas9 clones and multinucleated cells in CRISPR/Cas9 OCRL KO clones. (B) The percentage of HK-2 cells with multiple nuclei is plotted (mean and individual points of three independent experiments; 69–394 cells per condition scored in each experiment). (C) Representative images of mononuclear cells in cells transfected with control siRNA and multinucleated cells in cells transfected with OCRL siRNA. (D) The percentage of cells with multiple nuclei is plotted (mean and individual points of three independent experiments; 111–481 cells per condition scored in each experiment). (E) Wheat germ agglutinin and DAPI were used to visualize cell borders and nuclei in control and LS fibroblast primary cultures. (F) The percentage of cells with multiple nuclei is plotted (mean and individual points in three independent experiments; 99–171 cells per condition scored in each experiment). (G) OCRL-depleted cells were transfected with cDNAs encoding GFP, or with siRNA-resistant GFP-tagged WT OCRL (OCRL) or LS-causing mutant OCRL-G304E (G304E). Efficient knockdown of OCRL (90%–95%) in these cells and heterologous expression of GFP-tagged OCRL and G304E (red asterisk) were confirmed by western blotting of cell lysates. (H) Representative expression patterns of each construct are shown (green). (I) The percentage of cells with multiple nuclei in cells treated with siRNA and expressing GFP, OCRL, and G304E is plotted (mean and individual points of three experiments for GFP and OCRL, two experiments for G304E; 11–146 cells per condition scored in each experiment). Scale bar (A, C, E, and G), 10 µm. *P<0.05, **P<0.01, ***P<0.001, and ****P<0.001 on the basis of a one-way ANOVA Tukey’s multiple comparisons test in (B), unpaired t test in (D) and (F), and two-way ANOVA Tukey’s multiple comparisons test in (I). cl, clone; Exp, experiment.