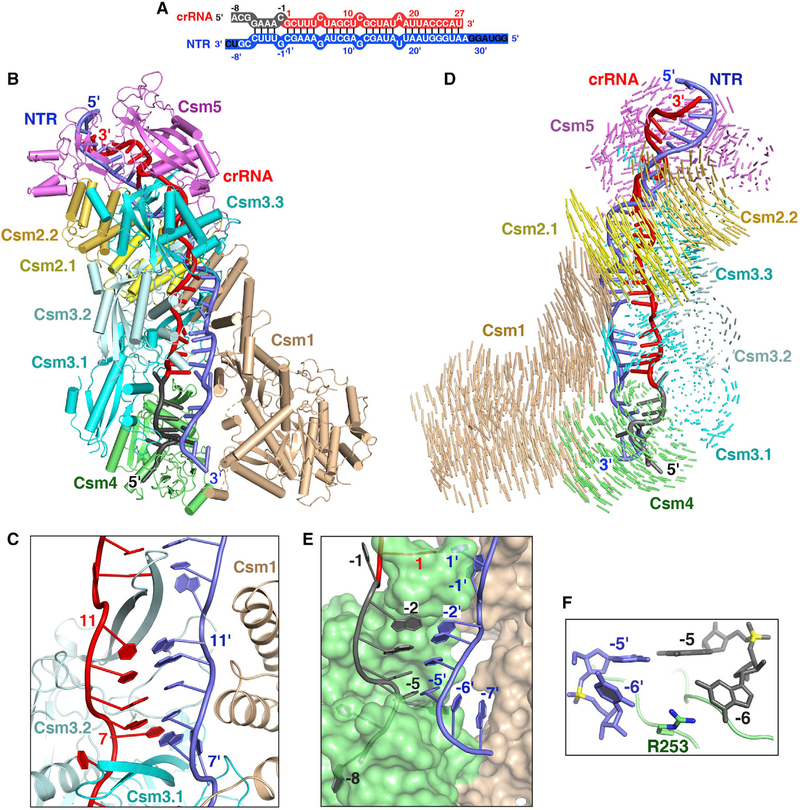
Figure 2. Cryo-EM Structure of the Csm-NTR Complex.
(A) Schematic representation of the crRNA-NTR duplex. The base pairs observed in the NTR-bound Csm complex are depicted by lines. The non-cognate target RNA is shown in blue. The ordered and disordered nucleotides of NTR are shown in white and black, respectively.
(B) Overall structure of the NTR-bound Csm complex.
(C) The β-thumb of each Csm3 subunit inserts into the crRNA-NTR duplex, leading to the periodical nucleotide displacement of both RNA strands.
(D) Structural comparison between Csm-crRNA binary complex and Csm-crRNA-NTR ternary complex showing the conformational change upon target RNA binding. Vector length correlates with the domain motion scale (color-coded as defined in Figure 1A).
(E) Nucleotides at positions (−2)′–(−5)′ in the 3′ anti-tag of NTR base pair with nucleotides (−2)–(−5) in the 5′ tag of the crRNA.
(F) The side-chain of Arg253 of Csm4 subunit inserts into the bases at position (−6) of crRNA and target RNA, preventing base pairing at this position.
See also Figures S2 and S3 and Table S2.