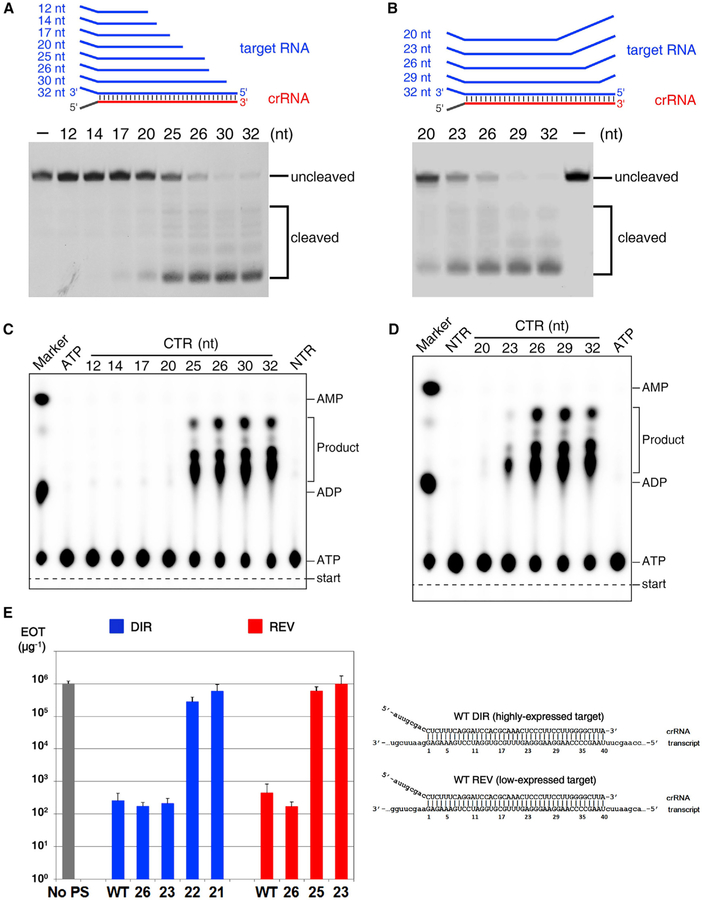
Figure 6. Determining the Minimal Length of crRNA Spacer Segment Required for DNase and cOA Synthetase Activities of the Csm Complex.
(A and B) DNA cleavage by various CTR-bound SthCsm complexes. The cognate target RNA is truncated from it 5′ end (A), and the target RNA contains mismatches from its 5′ end (B).
(C) cOAs synthesis assay with truncated cognate target RNA shown in (A).
(D) cOAs synthesis assay with the mismatch-containing cognate target RNA as shown in (B).
(E) Efficiency of transformation (EOT) of III-A+ T. thermophilus cells with plasmids carrying protospacers fully matched to (“WT”) or with multiple mismatches with the 3′ end of crRNA. Numbers indicate the length of crRNA-target duplex. Blue bars correspond to plasmids with protospacer cloned in direct orientation, which results in high level of transcripts targeted by crRNA, red bars correspond to plasmids with reverse protospacer orientation which results in reduced protospacer transcript levels. The gray bar indicates EOT with control plasmid without protospacer (“No PS”). The structures of crRNA-target duplexes for fully matching protospacers in both orientations are shown on the right. Mean values obtained from 3 independent experiments and SDs are shown.
See also Figure S6.