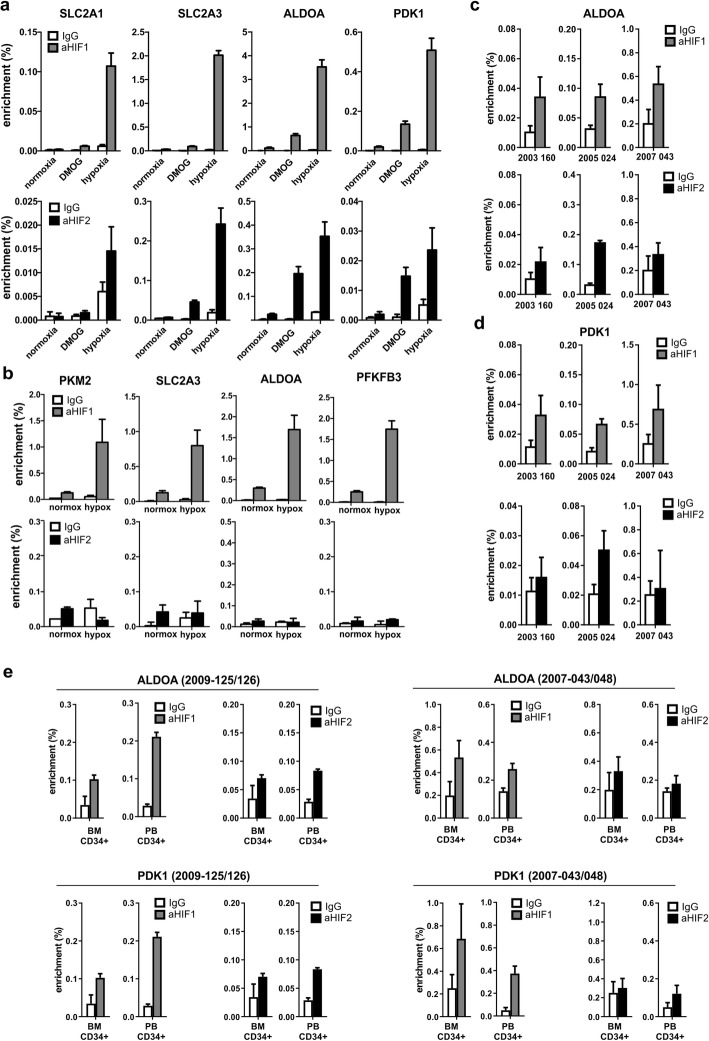
Fig. 2.
Validation of identified HIF1 and HIF2-bound loci in endogenous ChIP-PCRs. a HIF1 and HIF2 occupied loci identified by ChIPseq were validated in K562 using antibodies against endogenous HIF1 and HIF2. b HIF1 is more efficiently stabilized under hypoxia compared to HIF2 in CB CD34+ cells. Numbers below x-axis indicate patient sample numbers. c–d endogenous HIF1 and HIF2 ChIP PCRs on representative loci in primary AML CD34+ cells. e Endogenous HIF1 and HIF2 ChIP PCRs on representative loci in primary AML CD34+ cells derived from BM or PB. Numbers above graphs indicate patient sample numbers, whereby 2009-125 is derived from BM and 2009-126 is derived from PB from the same patient; 2007-043 is derived from BM and 2007-047 is derived from PB from the same patient