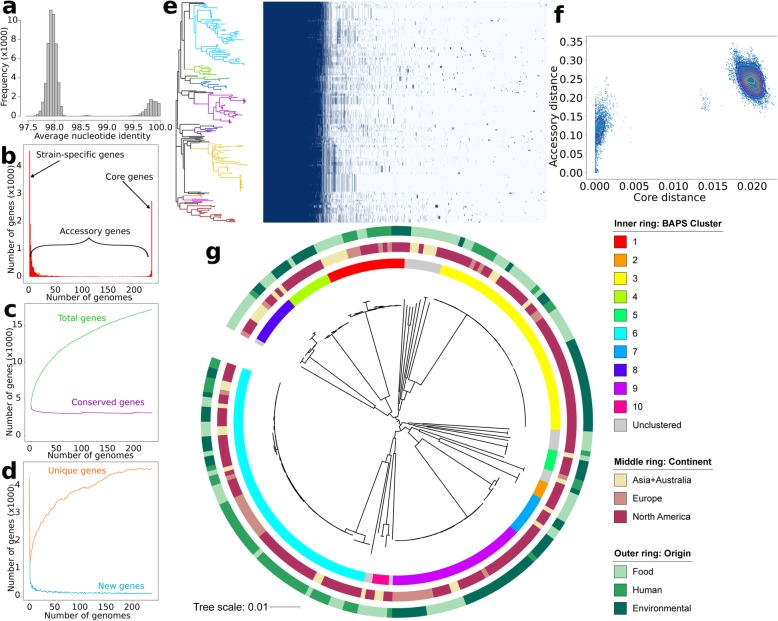
Fig. 1.
Pan-genome structure and phylogeny of C. sakazakii. a Distribution of pairwise ANI values. b The number of unique genes that are shared by any given number of genomes or unique to a single genome. Numerical values for each gene category are shown in Additional file 6: Table S3. c The size of the core genome (purple line) and pan-genome (green line) as more genomes are added. The list of core genes is listed in Additional file 7: Table S4. d The number of unique genes, i.e., genes unique to individual strains (orange line) and new genes, i.e., genes not found in the previously compared genomes (light blue line) as more genomes are added. e Gene presence-absence matrix showing the distribution of genes present in each genome. Each row corresponds to a branch on the tree. Each column represents an orthologous gene family. Dark blue blocks represent the presence of a gene, while light blue blocks represent the absence of a gene. The phylogeny reflects clustering based on presence or absence of accessory genes. The colors on the tip of each branch reflect the BAPS clustering. f Contour plots of pairwise distances between genomes in terms of their core genome divergence (measured by SNP density distance across the core genome) and the difference in their accessory genomes (measured by the Jaccard distance based on the variation in the gene content of their sequences) calculated using popPUNK [24]. g The midpoint-rooted maximum likelihood phylogenetic tree was calculated using sequence variation in the core genome alignment. Outer rings show the BAPS cluster, geographical origin, and ecological source. Scale bar represents nucleotide substitutions per site