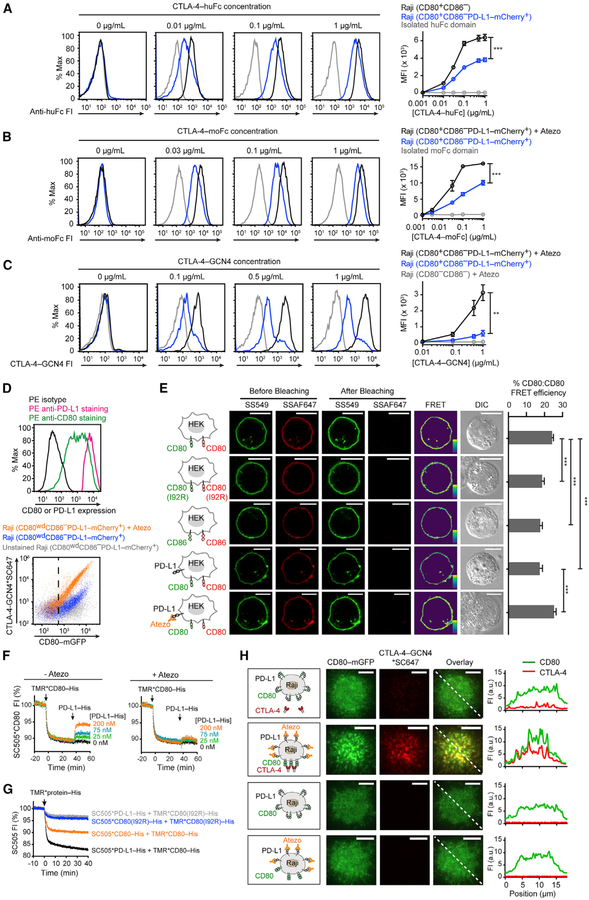
Figure 4. Cis-PD-L1 Inhibits CD80:CTLA-4 Interaction through Disrupting CD80 Homodimers.
(A) Representative flow-cytometry histograms of CTLA-4-huFc staining of the indicated types of Raji cells. Bound CTLA-4-huFc was labeled by AF647 anti-human IgG Fc, the MFI of which was plotted against (CTLA-4-huFc). Shown in gray are Raji (CD80+CD86−) cells stained by isolated huFc domain. Means ± SEM, n ≥ 3.
(B) Representative flow-cytometry histograms of CTLA-4-moFc staining of Raji (CD80+CD86−PD-L1-mCherry+) cells with or without atezolizumab (Atezo) (20 μg/mL). Bound moFc was labeled by AF647 anti-mouse IgG Fc, the MFI of which was plotted against (CTLA-4-moFc). Shown in gray are atezolizumab-treated Raji (CD80+CD86−PD-L1-mCherry+) cells stained by isolated moFc domain. Means ± SEM, n ≥ 3.
(C) Representative flow-cytometry histograms of CTLA-4-GCN4*SC647 staining of Raji (CD80+CD86−PD-L1-mCherry+) cells with or without atezolizumab and of Raji (CD80−CD86−) cells with atezolizumab. MFI of SC647 was plotted against the input concentration (means ± SEM, n ≥ 3).
(D) At the top are flow-cytometry histograms showing both PD-L1 and CD80 amounts on a population of Raji (CD80wdCD86−PD-L1-mCherry+) cells with tight PD-L1 expression and a wide range of CD80 expression. The cells were stained with either phycoerythrin (PE) anti-CD80, PE anti-PD-L1, or PE isotype, and the 3 histograms overlaid. On the bottom is a flow-cytometry dot plot showing CTLA-4-GCN4*SC647 staining of Raji (CD80wdCD86−PD-L1-mCherry+) cells with or without atezolizumab. Gray dots correspond to control signals of unstained cells. CD80+ cells were gated by the vertical dash line, determined by the mGFP signal of parental Raji (CD80−CD86−) cells.
(E) A FRET assay probing CD80:CD80 homodimerization on cell membranes. In the first row, the leftmost cartoon depicts a HEK293T cell expressing SNAP-CD80, with a subpopulation labeled with SS549 (donor) and the rest labeled with SSAF647 (acceptor). On the immediate right are pre- and post-bleaching confocal images of a representative cell. Further on the right is the calculated pseudo-color FRET efficiency image (yellow to purple spectrum denotes strong to weak FRET) and the DIC image. The second and third rows are the same as the first row except replacing SNAP-CD80 with SNAP-CD80 (I92R) or with SNAP-CD86. The fourth row is the same as the first row except with co-expressed unlabeled PD-L1. The fifth row is the same as fourth row except in the presence of atezolizumab. The bar graph summarizes the FRET efficiencies as mean ± SEM, n > 22 cells from 3 independent experiments. Scale bars, 10 μm.
(F) An LUV FRET assay for probing CD80:CD80 homodimerization and PD-L1 effects. Shown is a representative time course of normalized FI of LUV-bound SC505*CD80-His, challenged by TMR*CD80-His and then by indicated concentrations of unlabeled PD-L1-His, with or without atezolizumab (Atezo) (20 μg/mL).
(G) An LUV FRET assay showing that a single point mutation in CD80 disrupts both CD80:CD80 homodimerization and PD-L1:CD80 heterodimerization. Each indicated SC505 (energy donor)-labeled protein was pre-coupled to DGS-NTA-Ni containing LUVs through its His-tag, and challenged with TMR (energy acceptor)-labeled proteins as indicated. Shown are representative time courses of 3 independent replicates.
(H) Representative TIRF images of Raji (CD80-mGFP+CD86−PD-L1-SNAP+) cells stained with CTLA-4-GCN4*SC647, at the indicated channels, under the depicted conditions. Rightmost are FI profiles along the dashed line at the overlaid images. Scale bars, 5 μm. See also Figures S1–S4.
Unpaired two-tailed Student’s t test: *p < 0.05, **p < 0.01, ***p < 0.001. See Table S3 for genotypes of cells related to this figure.