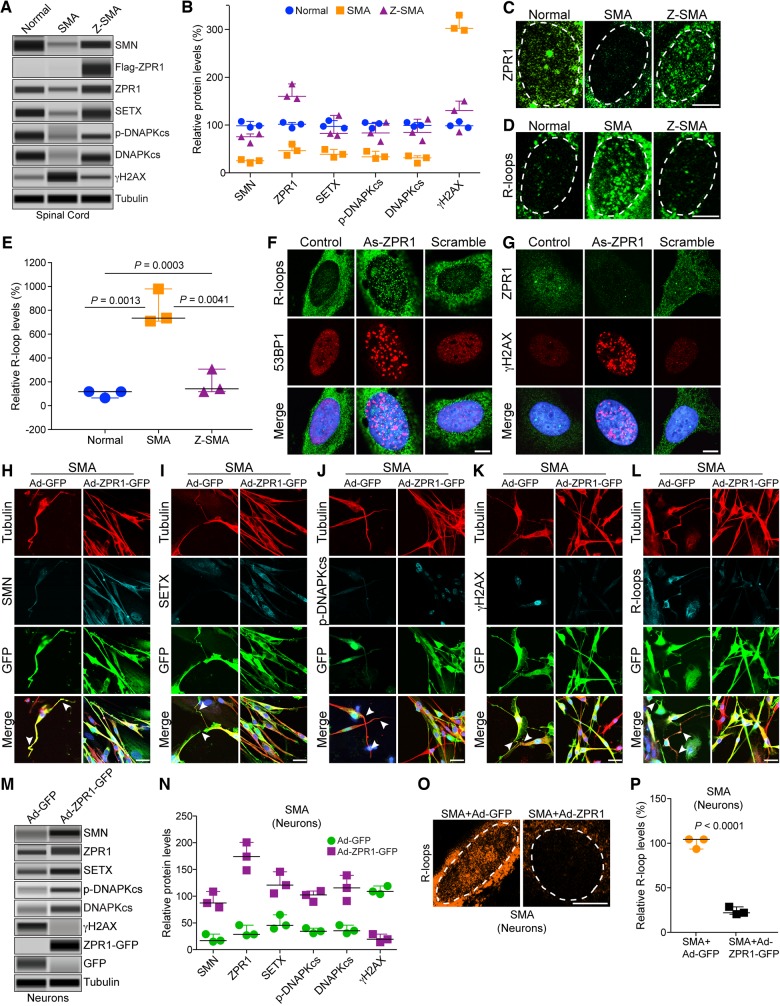
Figure 5.
ZPR1 rescues molecular defects and DNA damage associated with SMA pathogenesis in SMA motor neurons and SMA mice. Protein extracts were prepared from the spinal cords isolated from 7-day-old normal, SMAΔ7 (SMA) and Z-SMA mice, and examined using an automated capillary-based western blot system. Representative capillary-blot images of proteins are shown (full-length blots are provided in Supplementary Fig. 4). (A) Immunoblot analysis of spinal cord protein extracts. (B) Quantitative immunoblot data are presented as a scatter plot with median, min and max (median, min, max) range shows relative change (%) in levels of SMN and DNA damage markers caused by in vivo ZPR1 overexpression in the spinal cords of mice, SMN [SMA (25.64, 20.65, 27.98) and Z-SMA (75.69, 62.65, 75.69)], SETX [SMA (38.95, 32.74, 49.21) and Z-SMA (82.32, 78.32, 120.33)], p-DNA-PKcs [SMA (33.65, 30.21, 45.36) and Z-SMA (83.65, 65.79, 105.96)], total DNA-PKcs [SMA (31.70, 20.69, 35.68) and Z-SMA (84.65, 67.32, 112.36)] and γH2AX [SMA (302.69, 298.63, 330.25) and Z-SMA (130.65, 85.63, 150.32)]. Statistical analysis (mean ± SEM, n = 3 mice/group) using t-test (unpaired, two-tailed) of proteins shows increase in ZPR1 (167.40 ± 9.55%, P = 0.0005) in Z-SMA compared to ZPR1 (47.99 ± 6.78%) levels in SMA mice results in increase of SMN (73.33 ± 5.61%, P = 0.0013) in Z-SMA compared to SMN (24.76 ± 2.16%) levels in SMA mice, SETX (93.66 ± 13.39%, P = 0.0199) in Z-SMA compared to SETX (40.30 ± 4.80%) levels in SMA mice, phospho-DNA-PKcs (p-DNAPKcs) (85.13 ± 11.62%, P = 0.0175) in Z-SMA compared to p-DNA-PKcs (36.41 ± 4.58%) levels in SMA mice and total DNA-PKcs (88.11 ± 13.12%, P = 0.0133) in Z-SMA compared to DNA-PKcs (29.36 ± 4.48%) levels in SMA mice. Analysis of DNA damage response marker shows decrease in γH2AX (122.20 ± 19.15%, P = 0.0009) in Z-SMA compared to γH2AX (310.5 ± 9.93%) levels in SMA mice suggesting the rescue of DNA damage in vivo by ZPR1 overexpression. (C and D) Primary spinal cord neurons were cultured from 7-day-old normal, SMA and Z-SMA mice. (C) Neurons were stained with ZPR1 and (D) R-loops (S9.6) antibodies and high magnification images of nuclei are presented. Dotted ellipses represent nuclei. Scale bar = 5.0 μm. (E) Quantitative analysis of relative levels of accumulation of nuclear R-loops in motor neurons from normal, SMA and Z-SMA mice is presented as a scatter plot with IQR (median, min, max), normal (118.0, 65.25, 118.7), SMA (734.90, 709.40, 978.9) and Z-SMA (141.8, 118.20, 306.90). Statistical analyses (mean ± SEM, n = 3 experiments, 50 neurons/experiment) using t-test (unpaired, two-tailed) of accumulation of R-loops in SMA (807.7 ± 85.90%, P = 0.0013) and normal neurons (106 ± 17.69%) shows large (∼8-fold) increase in accumulation of R-loops in SMA compared to normal mice. Comparison of R-loop accumulation between SMA (807.7 ± 85.90) and Z-SMA (189.0 ± 59.36) using t-test (P = 0.0041) and comparison between normal, SMA and Z-SMA using ANOVA (P = 0.0003) shows statistically significant decrease in in vivo R-loop accumulation by ZPR1 overexpression in Z-SMA mice. (F and G) HeLa cells were transfected with mock (Control), antisense ZPR1 oligonucleotide (As-ZPR1) and scrambled oligonucleotides (Scramble) (100 nM). (F) Knockdown of ZPR1 results in accumulation of R-loops (green) in the nucleus and formation 53BP1 foci (red) suggesting DNA double-strand breaks (DSBs). (G) ZPR1 (green) knockdown results in loss ZPR1 nuclear foci and causes accumulation of γH2AX foci (red) suggesting activation of DNA damage response in response to DNA damage caused by ZPR1 deficiency. Scale bar = 5 μm. (H–L) Cultured primary spinal cord neurons from SMA mice were infected with adenovirus (100 MOI) expressing green fluorescent protein (GFP) (Ad-GFP) and ZPR1-GFP fusion protein (Ad-ZPR1-GFP) and stained with antibodies against neuron-specific β-tubulin-III (red), SMN, SETX, p-DNA-PKcs, R-loops and γH2AX, and immunofluorescence was examined by confocal microscopy. GFP and ZPR1-GFP (green) were detected by GFP fluorescence. Axonal defects include retraction, bending, folding of axons (arrowheads) that indicate degeneration of SMN-deficient neurons. (H) Staining of neurons with SMN (cyan) and β-tubulin (red), (I) SETX (cyan) and β-tubulin (red), (J) p-DNA-PKcs (cyan) and β-tubulin (red), (K) γH2AX (cyan) and β-tubulin (red) and (L) R-loops (cyan) and β-tubulin (red). SMA neurons with ZPR1 ectopic expression (Ad-ZPR1-GFP panels) show reduction in neuron degenerative features. Nuclei were stained with DAPI (blue). Scale bar = 25 μm. Enlarged images of merged panels (H–L) are included in Supplementary Fig. 5 to show features of axonal degeneration such as loosening, bending retraction and ballooning in SMA neurons. (M) Immunoblot analysis of in vitro cultured motor neurons from SMA mice expressing GFP (Ad-GFP) and ZPR1-GFP (Ad-ZPR1-GFP) for changes in levels of SMN and DNA damage markers, SETX, p-DNA-PKcs, total DNA-PKcs and γH2AX. (N) Quantitative immunoblot data are presented as a scatter plot with median, min and max range shows relative change (%) in levels of SMN and DNA damage markers caused by in vitro ZPR1 overexpression in cultured motor neurons from SMA mice, SMN [SMA+GFP (17.06, 15.02, 29.0) and SMA+ZPR1-GFP (87.32, 79.06, 108.63)], SETX [SMA+GFP (45.62, 39.52, 65.32) and SMA+ZPR1-GFP (120.69, 105.36, 145.63)], p-DNA-PKcs [SMA+GFP (34.56, 30.65, 40.31) and SMA+ZPR1-GFP (102.36, 89.65, 109.65)], total DNA-PKcs [SMA+GFP (35.62, 30.65, 45.98) and SMA+ZPR1-GFP (115.47, 91.20, 138.97)] and γH2AX [SMA+GFP (108.90, 104.04, 118.99) and SMA+ZPR1-GFP (19.45, 14.13, 28.96)]. Statistical analysis using unpaired t-test of quantitative data (mean ± SEM, n = 3 mice/group) from spinal cord neuron immunoblots shows increase in ZPR1 levels (5.14 ± 0.44, P = 0.0010)-fold results in marked increase in levels of SMN (4.50 ± 0.43, P = 0.0019)-fold, SETX (2.47 ± 0.23, P = 0.0075)-fold, p-DNA-PKcs (2.85 ± 0.16, P = 0.0006)-fold and total DNA-PKcs (3.07 ± 0.36, P = 0.0059)-fold leading to decrease in γH2AX levels (5.04 ± 0.03, P = 0.0002)-fold. These data suggest the rescue of DNA damage in SMA spinal cord neurons (full-length blots are provided in Supplementary Fig. 6). (O) Enlarged images of nuclei of neurons stained with S9.6 antibody (R-loops, pseudocoloured orange) from SMA+Ad-GFP (SMA) and SMA+Ad-ZPR1-GFP (SMA+ZPR1) groups of SMA neurons. (P) Quantitative and statistical analysis of R-loop accumulation in cultured SMA motor neurons, SMA+GFP (103.5, 92.80, 103.7) and SMA+ZPR1-GFP (21.36, 19.65, 27.89) shows reduced accumulation (22.97 ± 2.51%, P < 0.0001) in neurons overexpressing ZPR1 (SMA+ZPR1-GFP) compared SMA+ GFP neurons.