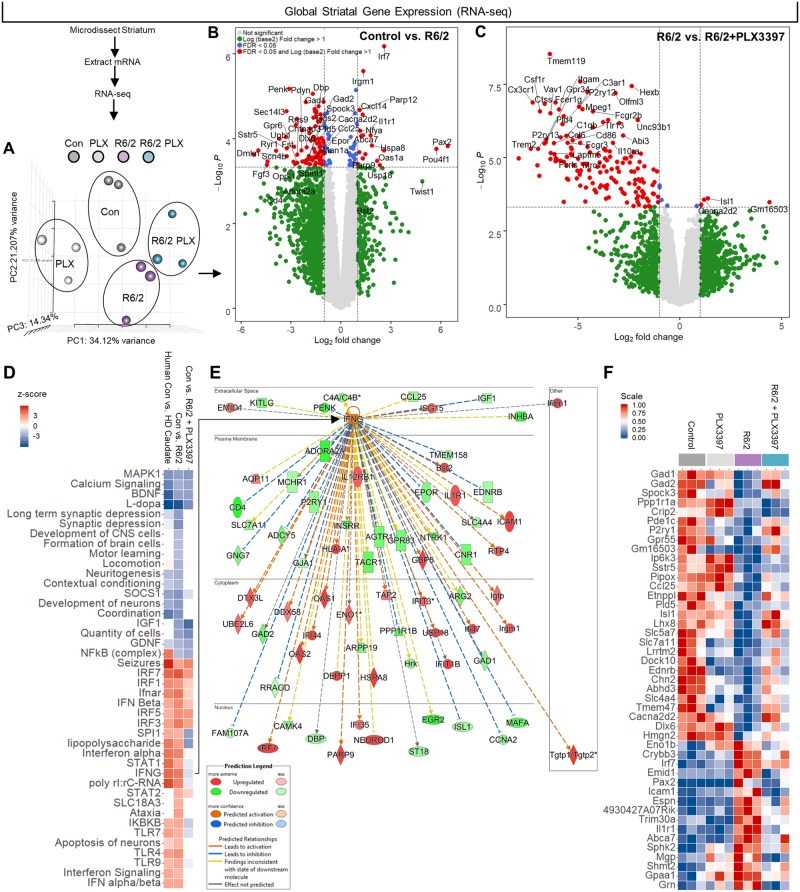
Figure 3.
RNA-seq and pathway analysis confirm lack of inflammatory transcriptional polarization in R6/2 striatum, except for a dysregulated interferon signature resembling the human Huntington’s disease caudate that is partially resolved following CSF1Ri. (A) PCA of striatal gene expression as determined by mRNA-seq confirmed similar non-overlapping clustering of control and R6/2 samples but a divergent effect of PLX3397 (n = 3/group). (B and C) Volcano plots of control versus R6/2 DEGs (B) confirmed an absence of inflammatory transcript upregulation in the R6/2 striatum and (C) a loss of microglial gene expression with PLX3397 (FDR ≤ 0.05; n = 3/group). (D) Pathway analysis (IPA) of 860 control versus R6/2 DEGs and 1380 control versus R6/2+PLX3397 DEGs (FDR ≤ 0.1; n = 3/group) revealed a distinct interferon signature and suppressed neuronal development and neuritogenesis pathways in the R6/2 striatum that closely resembled the human Huntington’s disease caudate, and which were partially resolved following PLX3397. Top significant predicted upstream regulators included HTT, IFNγ, l-DOPA, IRF7, and TRIM24, with associated activation z-scores of 2.151 (P < 8.31 × 10−22), 3.727 (P < 1.01 × 10−15), −5.654 (P < 1.19 × 10−20), 4.589 (P < 7.47 × 10−13), and −3.970 (P < 3.58 × 10−12), respectively. (E) IPA predicted IFNγ as the major cytokine upstream regulator in the R6/2 striatum, signalling that, along with IFNα, is no longer enriched in R6/2+PLX3397 striatum. (F) A heat map of control versus R6/2 DEGs (FDR < 0.1) that were reversed with treatment by unadjusted P-value (P < 0.05) to indicate potential mediators of beneficial PLX3397 effects (n = 3/group). All RPKM values can be searched and visualized at http://rnaseq.mind.uci.edu/green/R62_PLX/gene_search.php.