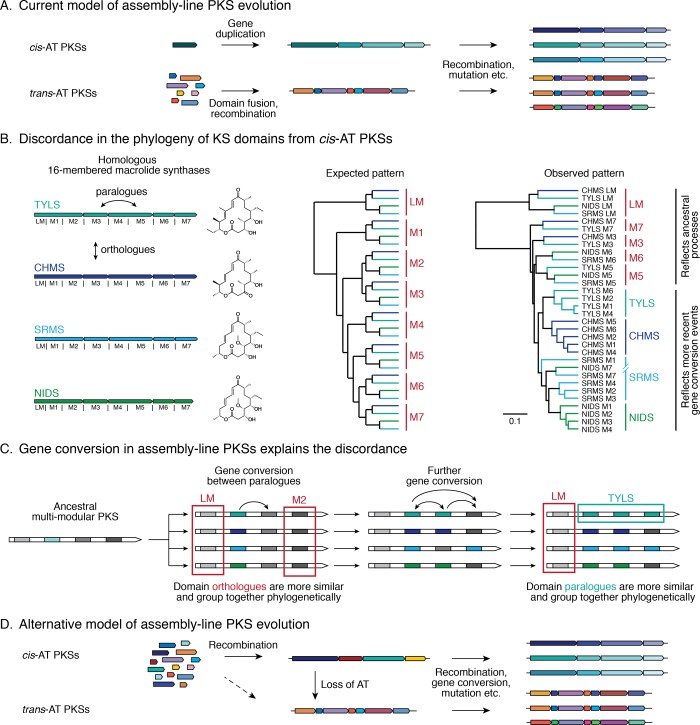
Figure 3.
Models of cis-AT and trans-AT PKS evolution. (A) It has been hypothesized that evolution of cis- versus trans-AT PKSs took distinct paths.21,32,34 However, this dichotomy has some discordances. It does not explain the absence of iterative trans-AT PKSs, the convergence toward strikingly similar architectures despite different evolutionary paths, the presence of AT domain vestiges in trans-AT modules,34 or (B) the inconsistency of the phylogenetic tree of cis-AT KS domains with this hypothesis.64 The last inconsistency is exemplified by KS domains from four homologous 16-membered macrolide synthases (left; TYLS, tylactone synthase; CHMS, chalcomycin synthase; SRMS, spiramycin synthase; NIDS, niddamycin synthase). Under the current model, their KS domains would be expected to form groups of orthologous domains (center). In fact, most KS domains are grouped with paralogues from the same PKS (right). Protein sequence alignment was performed with ClustalOmega,84 and the dendrogram was constructed using UPGMA hierarchical clustering. (C) The discordance in KS sequence alignment is a result of concerted evolution and can be explained by gene conversion events between KS domains.64,82 Gene conversion leads to high sequence similarity between paralogous domains, causing them to cluster closer to each other than to their orthologues (e.g., teal square). Because gene conversion need not affect all domains within a PKS (e.g., red square), some of them maintain a phylogenetic pattern reflecting ancestral events that had led to the separation of homologous assembly-line PKSs. (D) An alternative model for assembly-line PKS evolution builds on the hypothesis that trans-AT PKSs evolved from cis-AT PKSs through loss of AT domains. In this model, the high sequence identity of KS domains in cis-AT PKSs would be explained by subsequent gene conversion events rather than ancestral gene duplications.