Figure 4.
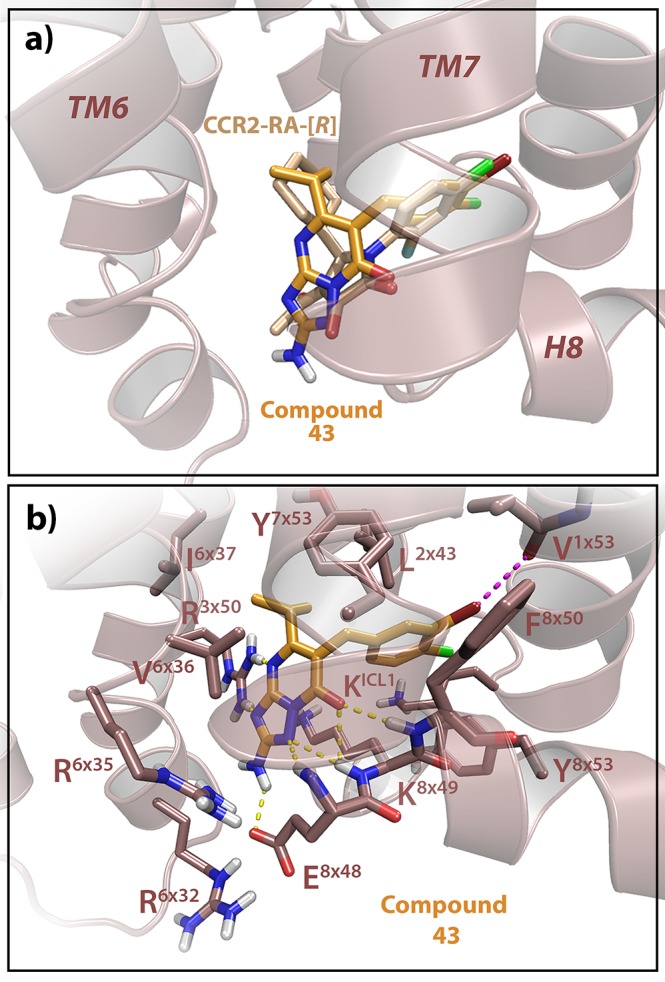
Proposed binding mode of 43 in hCCR2b. (a) Overlay of 43 with the CCR2 intracellular ligand CCR2-RA-[R], showing that 43 interacts in a similar manner as CCR2-RA-[R]. (b) Docking of 43, displaying the interactions with CCR2. The amino group in R2 makes an extra hydrogen-bond interaction with E8×48, while the bromine group in R1 makes an extra halogen bond with the backbone of V1×53, which might contribute to the improved affinity of 43 to this receptor. Model of hCCR2 is based on the crystal structure of CCR2 (PDB code 5T1A),15 and amino acid residues are labeled according to their structure-based Ballesteros–Weinstein numbers.25
