FIG 1.
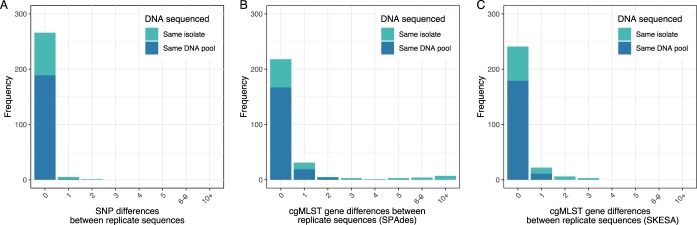
Observed differences using SNP typing (panel A) and hash-cgMLST based on SPAdes (panel B) and SKESA (panel C) assemblies in 272 replicate sequence pairs. With perfect sequencing, no variants would be expected between pairs of sequences from the same isolate. Pairs of sequences known to have been obtained from the same pool of DNA are shown in dark blue. Where information was unavailable on whether the same pool of DNA was used or a fresh DNA extract was made from the same isolate, this is shown in light blue.