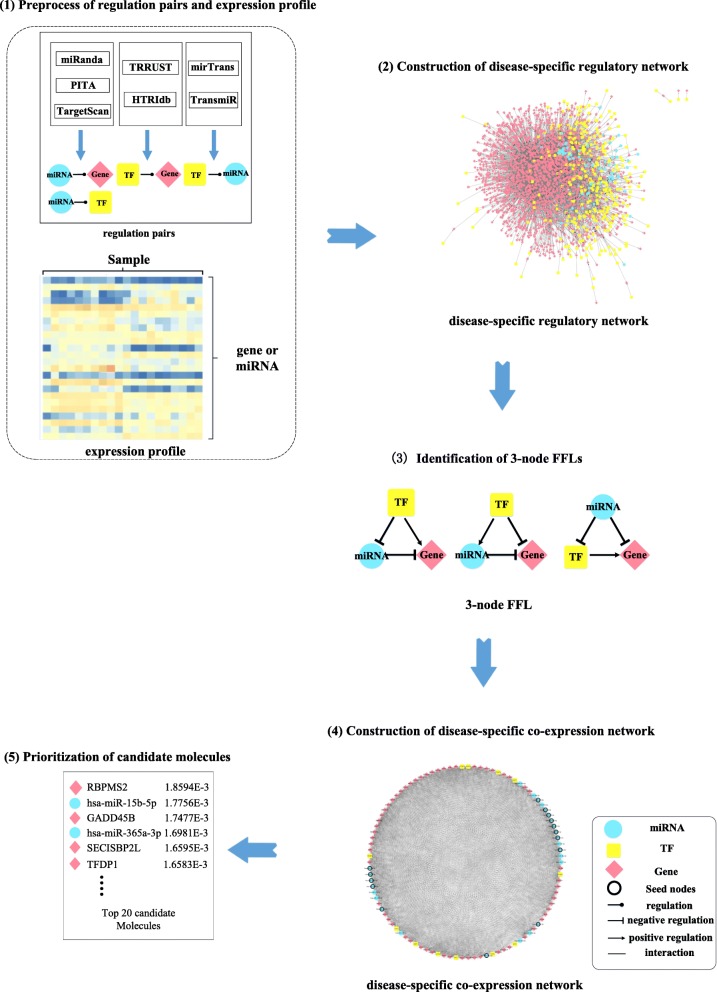
Fig. 1.
Flowchart of the proposed computational framework. (1) Preprocessing of regulation pairs and expression profiles (2) Construction of disease-specific regulatory networks (3) Identification of 3-node FFLs (4) Construction of disease-specific co-expression networks (5) Prioritization of candidate molecules