Table 1.
CRISPR-based barcode designs. From left to right, name and model organism, properties and schematic of various barcode designs available to date. Different targets are represented by different colours. Barcode properties: synthetic barcodes are transgenes artificially integrated into the host genome; endogenous barcodes are naturally present in the genomic DNA. Arrays involve multiple targets in tandem. Dispersed barcodes involve multiple targets present far apart in the genome. For each barcode, the number of identical (distinguishable or indistinguishable) or different targets is specified. Barcodes can be retrieved by sequencing (involves tissue dissociation, cell isolation and nucleic acid tagging) or by imaging (involves the hybridization of predefined fluorescent RNA probes to the edited or unedited barcode RNA). gRNAs are ubiquitously expressed in all systems except for MEMOIR, which relies on a Wnt-inducible gRNA. ID: molecular identifier.
| name (model) | barcode properties | scheme | ref. |
|---|---|---|---|
| GESTALT (in vitro and zebrafish) |
synthetic array 10 different targets sequencing retrieval |
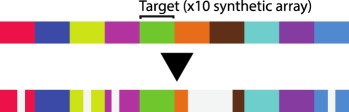 |
[11,12] |
| Scartrace (zebrafish) |
synthetic array 8 indistinguishable GFPs sequencing retrieval |
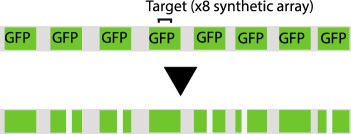 |
[13] |
| LINNAEUS (in vitro and zebrafish) |
synthetic dispersed 32 indistinguishable RFPs sequencing retrieval |
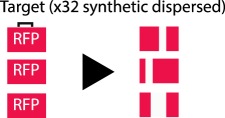 |
[14] |
| MARC1 (mouse) |
synthetic dispersed 60 different self-targeting gRNAs sequencing retrieval |
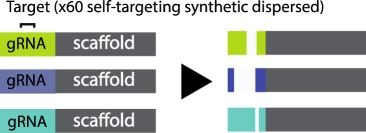 |
[15] |
| SEQuoia (Drosophila) |
synthetic array 32 distinguishable variants of one target sequencing retrieval |
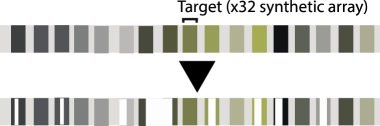 |
[16] |
| molecular recorder (mouse) |
synthetic array 3 different targets distinguishable (ID) multicopy integrations sequencing retrieval |
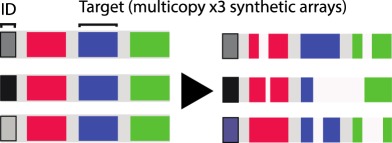 |
[17] |
| Cotterell & Sharpe (mammalian cells and zebrafish) |
multiple endogenous arrays 10 different targets/array sequencing retrieval |
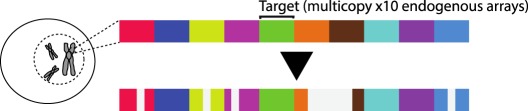 |
[18] |
| Byungjin Hwang et al. (mammalian cells) |
endogenous L1 repeats >200 distinguishable targets single gRNA/Cas9 base editor variant sequencing retrieval |
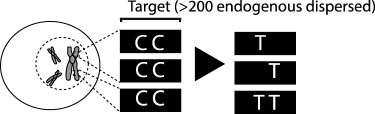 |
[19] |
| MEMOIR (mammalian cells) |
synthetic array 10 indistinguishable targets distinguishable (ID) multicopy integrations inducible gRNA imaging retrieval (unedited versus edited state) |
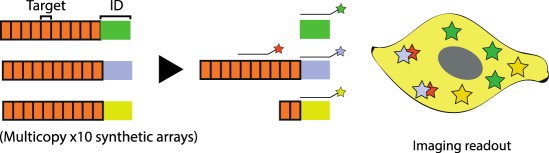 |
[20] |
