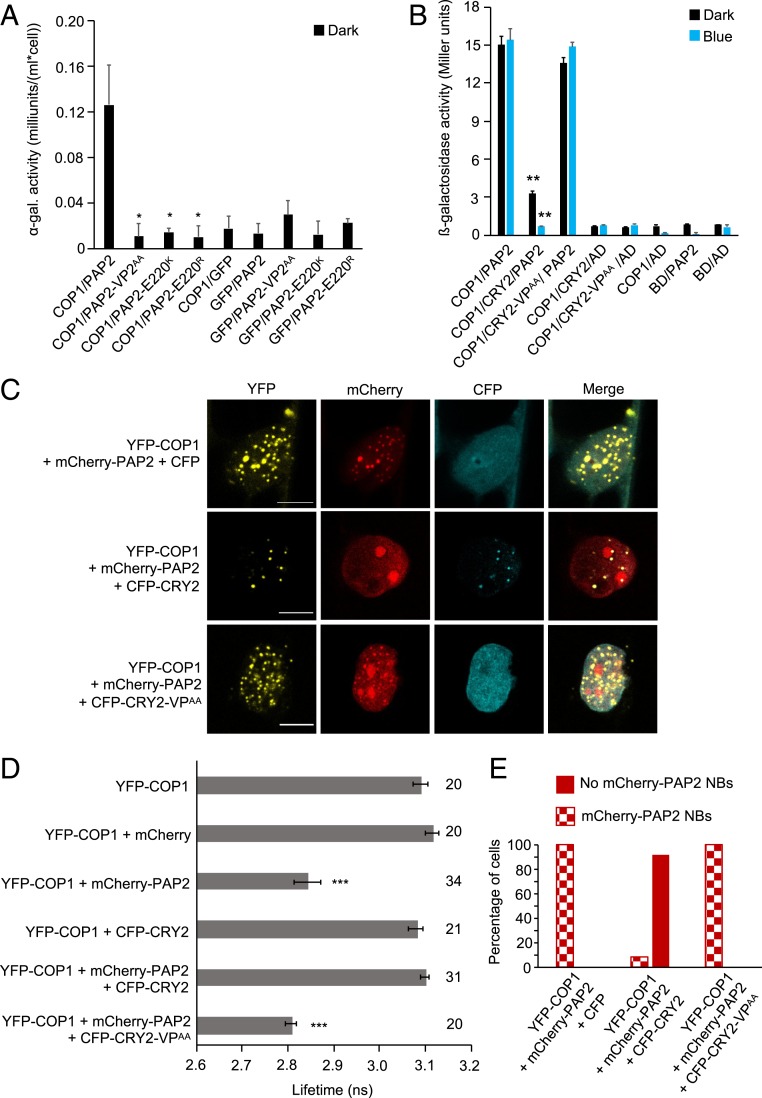
Fig. 5.
CRY2 competes with PAP2 for interaction with COP1. (A) Y2H assays with COP1 as bait and PAP2, PAP2-VP2AA, PAP2-E220K, or PAP2-E220R as prey. GFP served as a negative control. ⍺-galactosidase activity was measured in dark-grown yeast cells coexpressing bait and prey proteins. The asterisk indicates significant difference in COP1-interaction strength of PAP2VP2AA, PAP2-E220K, or PAP2-E220R when used as prey proteins compared to when full-length PAP2 was used (*P < 0.05). (B) Y3H assay with COP1 as bait and PAP2 as prey in the presence or absence of CRY2 or CRY2-VPAA. β-galactosidase activity was measured in cotransformed yeast cells after they were grown for 4 d in darkness (dark) or for 4 d in darkness followed by exposure to blue light (50 µmol m-2 s-1) (blue) for 4 h. Error bars indicate SEM. Asterisks indicate a significant difference in the interaction between COP1 and PAP2 in the presence of CRY2 compared with the absence of CRY2, under blue light (**P < 0.01). (C) Representative confocal images of leek cells coexpressing the indicated proteins. The indicated channels were subsequently merged to show colocalization. (Scale bars: 10 µm.) (D) Lifetime of donor (YFP-COP1) fluorophore measured by FRET-FLIM inside the nuclear bodies of leek cells after particle bombardment. The number adjacent to each bar represents the number of cells measured. Error bars represent SEM. Asterisks indicate a significant difference in lifetime compared with YFP-COP1 (***P < 0.001). The lifetime of YFP-COP1 was measured without exciting the CFP-tagged proteins. (E) Percentage of cells in which mCherry-PAP2 NBs were formed when YFP-COP1 and mCherry-PAP2 were coexpressed with CFP, CFP-CRY2, or CFP-CRY2-VPAA. Sixty cells were evaluated for each combination.