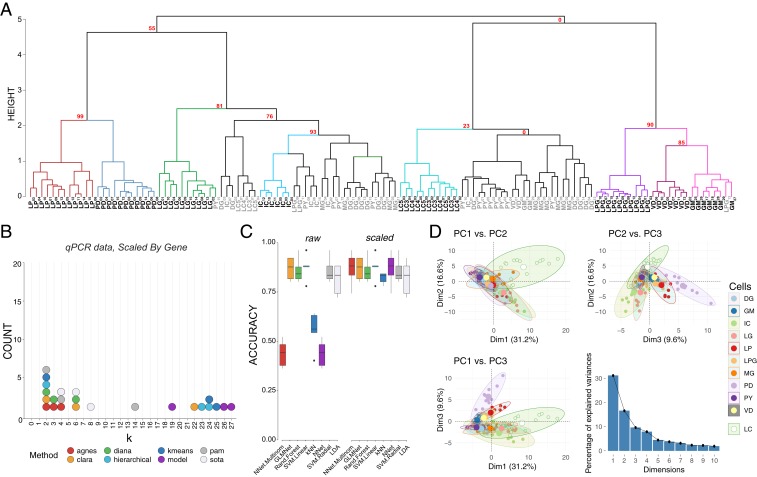
Fig. 4.
Post hoc recapitulation of cell identity via qRT-PCR expression with hierarchical clustering and sML algorithms. (A) Hierarchical clustering of cell type with correlation as the distance metric and Ward.D2 as the clustering method for data centered and scaled across genes. AU P values for a given node are noted in red. Each node that has >80% support by AU P value is color coded, and cell types that form a largely coherent group are noted in bold. Cells that do not appear to cluster by type are noted in gray. Cells are identified by type and a subscript that denotes a unique sample identifier. (B) Dotplot of the top 3 predicted number of clusters based on 8 different prediction algorithms. None of these methods correctly predicted 11 distinct clusters that would represent the 11 different cell types in this assay. (C) Accuracy of cell-type prediction using 8 different methods of sML for each of the datasets. Box and whisker plots show efficacy of each method across 5 cross-validation folds. (D) PCA for qRT-PCR data. Pairwise comparisons of PC1, PC2, and PC3 are shown in each panel as in Fig. 3. PC1 accounted for 31.2% of the variance, PC2 accounted for 16.6%, and PC3 accounted for 9.6% of the total variance across samples. A scree plot shows the amount of variance explained by PCs 1 through 10.