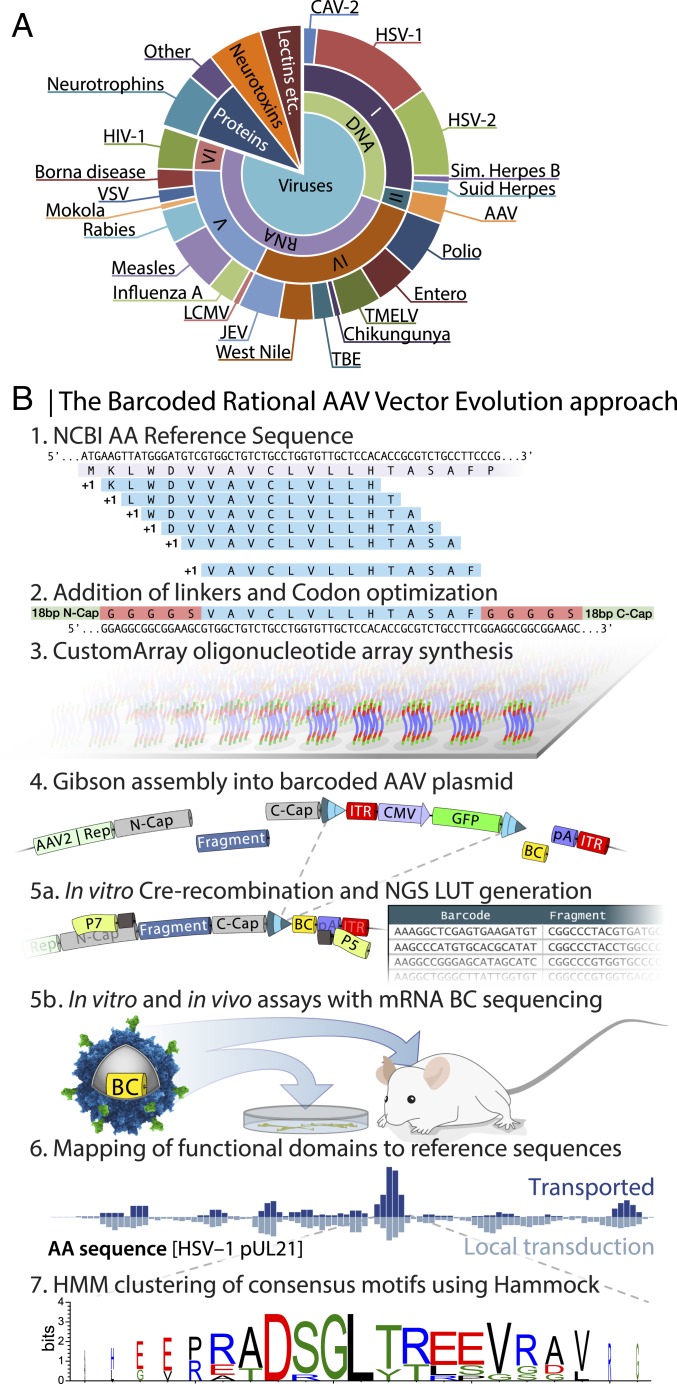
Fig. 1.
Barcoded rational AAV vector evolution screening approach. (A) Pie chart displaying the selected 131 proteins with documented affinity to synapses. (B) Schematics of the BRAVE procedure. 1: NCBI reference amino acid sequences were computationally digested into 14-aa (or 22-aa) -long polypeptides with a 1-aa shifting sliding window. 2: Three alternative linkers were added to the 14-aa polypeptides, for example, a rigid linker with 5 alanine residues (14aaA5). 3: Codon-optimized sequences (92,358) were synthesized in parallel on an oligonucleotide array. 4: The pool of oligonucleotides was assembled into an AAV production backbone with cis-acting AAV2 Rep/Cap and ITR-flanking CMV-GFP. A 20-bp random molecular barcode (BC) was simultaneously inserted into the 3′ UTR of the GFP gene. 5a: Using a fraction of the plasmid preparation, exposed to Cre-recombinase in vitro, a look-up table was generated linking the BC to the peptide. 5b: Using the same plasmid pool from step 4, an AAV batch was produced (MNMlib), and multiple parallel screening experiments were performed both in vitro and in vivo followed by BC sequencing from mRNA. 6: Through the combination of the sequenced barcodes and the LUT, transduction efficacy can be mapped back to each of the original 131 proteins, here exemplified by the aa sequence of HSV-1 pUL21. The height of each bar represents the relative abundance of that amino acid, normalized to the total library and summed from all peptides covering the region. 7: Consensus motifs can be determined using the Hammock, hidden Markov model-based clustering approach.